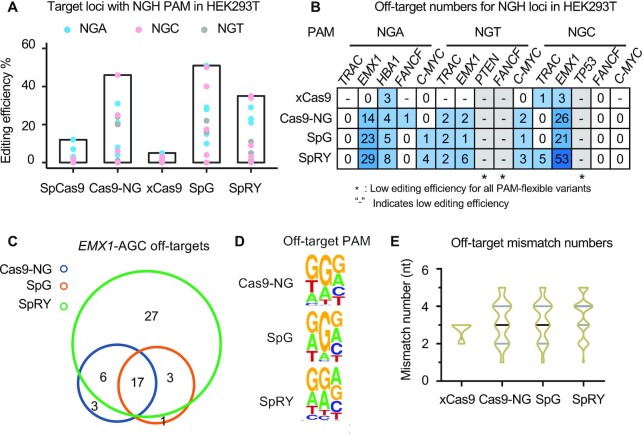
Figure 2.
Comparison of SpCas9, Cas9-NG, xCas9, SpG and SpRY at NGH loci. (A) Editing efficiency for SpCas9 and indicated PAM-flexible variants at 15 NGH (NGA, NGC or NGT) loci in HEK293T cells detected by PEM-seq. (B) Off-target numbers at indicated NGH loci detected by PEM-seq. ‘-’ indicates nearly no editing activity. * indicates low editing efficiency for all variants. PAM and locus information are marked above. (C) Venn diagram for off-targets of Cas9-NG, SpG and SpRY at EMX1-AGC locus detected by PEM-seq. (D) Consensus analysis by weblogo for PAM sequence for Cas9-NG, SpG and SpRY off-targets for all NGH loci detected by PEM-seq. (E) Statistics of sgRNA mismatch numbers between off-target and on target for all 15 NGH loci for indicated SpCas9 variants detected by PEM-seq.