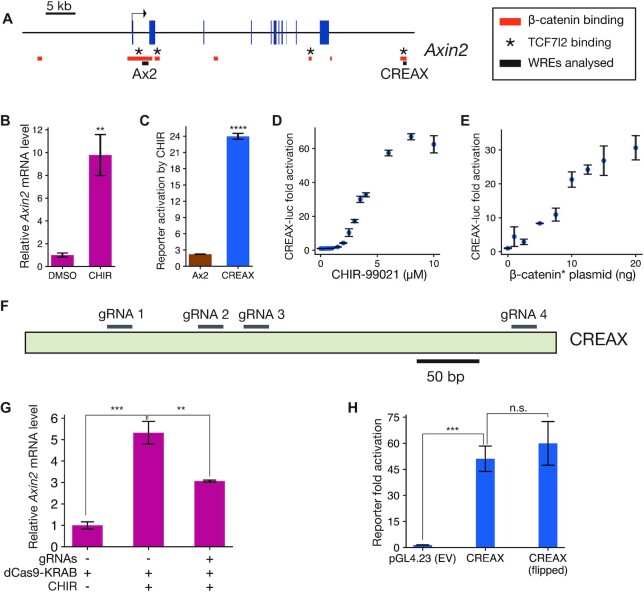
Figure 1.
Characterisation of CREAX (CDX Regulated Enhancer of Axin2), a highly Wnt-responsive enhancer near Axin2. (A) Position of CREAX (CDX Regulated Enhancer of Axin2) relative to the Axin2 gene locus. Blue boxes represent exons and red bars are regions bound by β-catenin in LS174T cells (Mokry et al., 2010). Known TCF7L2-bound regions in HEK293T cells are denoted by asterisks (Doumpas et al., 2018). Black bars denote regions cloned to generate the Ax2 and CREAX reporters. Positions are shown with respect to the hg18 assembly. (B) RT-qPCR measurement of Axin2 transcript levels showing an increase upon treatment with CHIR-99021 (CHIR) in HEK293T cells. (C) Activity levels of luciferase reporters of the previously identified Ax2 promoter-proximal WRE (Jho et al., 2002) and CREAX with CHIR treatment shows that CREAX is more sensitive to Wnt signalling. (D) CREAX-luciferase reporter activity increases in a dose-dependent manner to CHIR concentration in HEK293T cells. (E) CREAX-luciferase reporter activity increases in a dose-dependent manner upon transfection of increasing amounts of a plasmid expressing stabilised β-catenin containing the S33Y mutation (β-catenin*). (F) Cartoon showing positions of guide RNAs used to recruit the dCas9-KRAB construct to the CREAX locus for CRISPRi experiments. (G) RT-qPCR results showing that the increase in Axin2 transcript levels upon CHIR treatment is suppressed by the recruitment of dCas9-KRAB to the CREAX locus. (H) Activation of the CREAX-luciferase reporter by β-catenin* is not affected by orientation, meeting the classical definition of an enhancer. In (B-E,G, and H), data are presented as mean ± SD from three replicates (N = 3) for each condition. *P< 0.05; **P< 0.005; ***P< 0.001; ****P < 0.0001; n.s. P > 0.05.