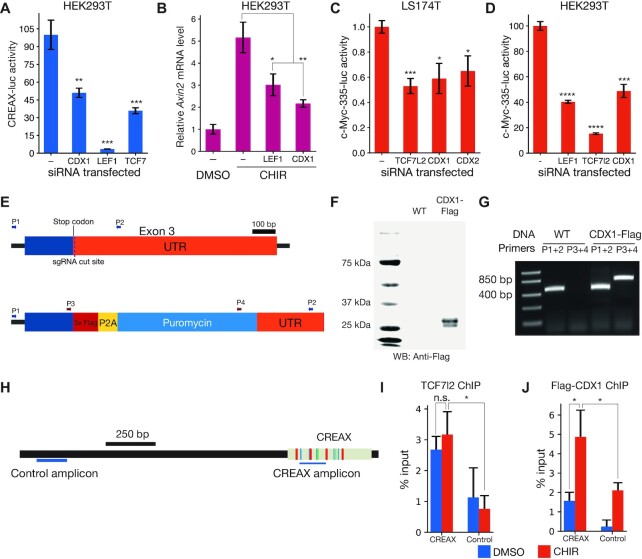
Figure 2.
CDX and TCF family members directly bind to and regulate the activity of CREAX and a distal enhancer of c-Myc. (A) RNAi-mediated depletion of CDX1, LEF1, or TCF7 reduces the activation of CREAX-luciferase by β-catenin* in HEK293T cells. Cells were transfected with CREAX-luc, β-catenin* and CDX1, LEF1, or TCF7 shRNA expression plasmids. (B) RNAi-mediated depletion of CDX1 or LEF1 reduces the increase in Axin2 mRNA levels caused by CHIR treatment in HEK293T cells. (C) Activity of a reporter of the c-Myc-335 enhancer previously shown to be bound by TCF7L2 and CDX2 in LS174T cells is reduced by RNAi against TCF7L2, CDX1 or CDX2. Cells were transfected with the reporter and plasmids expressing shRNA targeting TCF7L2, CDX1 or CDX2. (D) CHIR-stimulated c-Myc-335 reporter activity is decreased by RNAi-mediated depletion of LEF1, TCF7l2 and CDX1 in HEK293T cells. (E) Cartoon showing a portion of the CDX1 gene locus in HEK293T cells edited to express C-terminally Flag-tagged CDX1. The different sets of primers used to validate the edits are shown with arrows. (F) Anti-Flag western blot showing the expression of Flag-tagged protein in the CDX1-flag cells. (G) PCR validation showing that the CDX1-flag cells are heterozygous for the CDX1-flag allele. (H) Cartoon showing positions of the amplicons used to measure the ChIP signals from the CREAX and control loci. (I) TCF7L2 ChIP-qPCR signals from the CREAX and control loci in Flag-CDX1 HEK293T cells with and without a 24 h CHIR treatment. ChIP signals were measured using primers shown in panel H and normalized to signal from input chromatin. (J) Flag ChIP-qPCR signals from CREAX and control loci in Flag-CDX1 HEK293T cells with and without a 24 hour CHIR treatment. In (A-D,I,J), data are shown as mean ± SD from three replicates (N = 3) for each condition. P-value comparisons are made with the control condition unless otherwise indicated. P< 0.05; **P< 0.005; ***P< 0.001; ****P < 0.0001; n.s. P > 0.05.