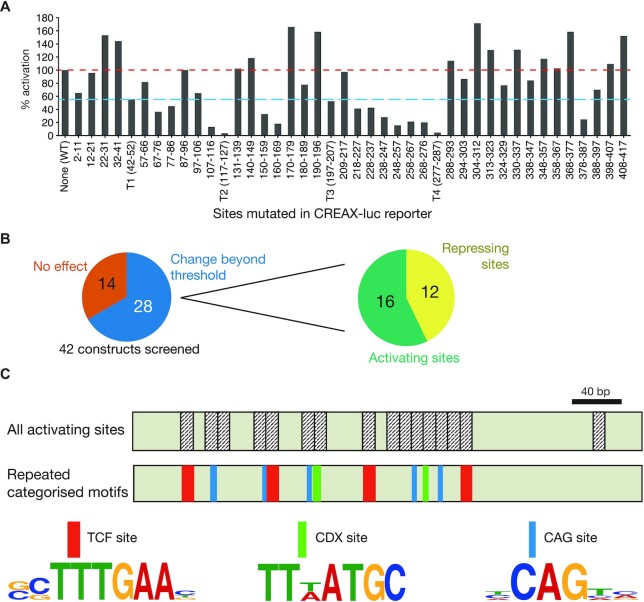
Figure 5.
Identification of multiple sets of recurring DNA motifs important for the activation of CREAX by Wnt signalling by a systematic mutagenesis screen. (A) Luciferase assay showing fold activation by β-catenin* of CREAX mutant constructs in HEK293T cells. Four TCF-binding sites were identified and mutated (labelled T1-T4), and the remaining nucleotides were mutated as indicated for non-overlapping screening mutagenesis. Mutations which decreased fold activation to at least 55% (blue line) or more than 100% of WT (red line) were considered hits from the screen. Bars represent a mean of two independent experiments (N = 2). (B) 28 of the 42 CREAX mutagenesis contructs showed significant changes in activation by β-catenin*. Of the 28, 16 showed reduced activity (suggesting that these regions play a role in enhancer activation) and 12 showed increased activity (suggesting a repressive effect). (C) The cartoon depicts the 420 bp CREAX fragment with the activating regions annotated. In addition to TCF binding sites (red), CREAX activity is regulated by multiple CDX binding sites (green) and CAG sites (teal), which are shown with their site logos.