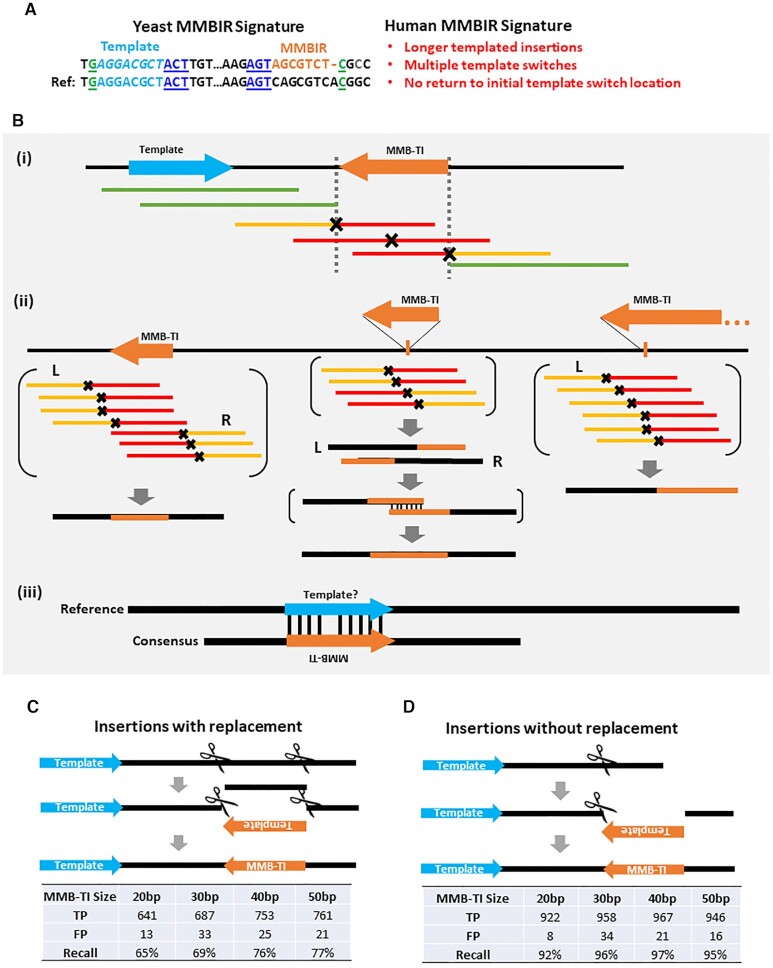
Figure 1.
MMBSearch is a novel tool to detect MMB-TI events in NGS reads. (A) Signatures of MMBIR from yeast and from human genomes that produce MMB-TIs. (Left) Top line: The outcome (signature) of MMB-TIs from studies in yeast (21) is an insertion (orange text) located in proximity and inverted orientation to its complementary template (light blue text) and flanked by microhomologies at the junctions (green and dark blue underlined text). Bottom line: The original sequence (Ref). (Right) Features of MMB-TIs in humans (red text) based on studies of CNVs in congenital diseases (8,10–12,16–20). (B) The MMBSearch tool identifies MMB-TIs based on the signatures described in (A): (i) reads containing MMB-TIs (red), which do not align to a reference genome are collected and re-aligned as half-reads. (ii) Reads where the first half (yellow) aligns while the second half (red) does not are collected and clustered by position of the aligned halves. The aligned read halves serve as anchors, and whole reads anchored on the same side (right – R or left – L) are used to create consensus sequences. If R and L consensuses overlap by position and are discordant (middle example), they are aligned to each other to create the full accurate consensus, which is compared back to the reference genome to identify the full MMB-TI sequence. (iii) The reverse-complement of the MMB-TI is aligned to the reference genome within 100 bp from the insertion to identify a possible template. (C) (top) Schematic for creation of synthetic insertions by replacing sequence with the reverse complement of a template from 80bp upstream (5′ of the insertion). (bottom) Number of true positives (TP), number of false positives (FP) and recall (TP/total) called by MMBSearch from processing of synthetic reads generated from Chromosome 17 with 993 synthetic insertions sized 20–50 bp. (D) (top) Schematic for creation of synthetic insertions by appending the reverse complement of a template from 80bp upstream (bottom). Analysis for the reads in top analyzed similarly to (C).