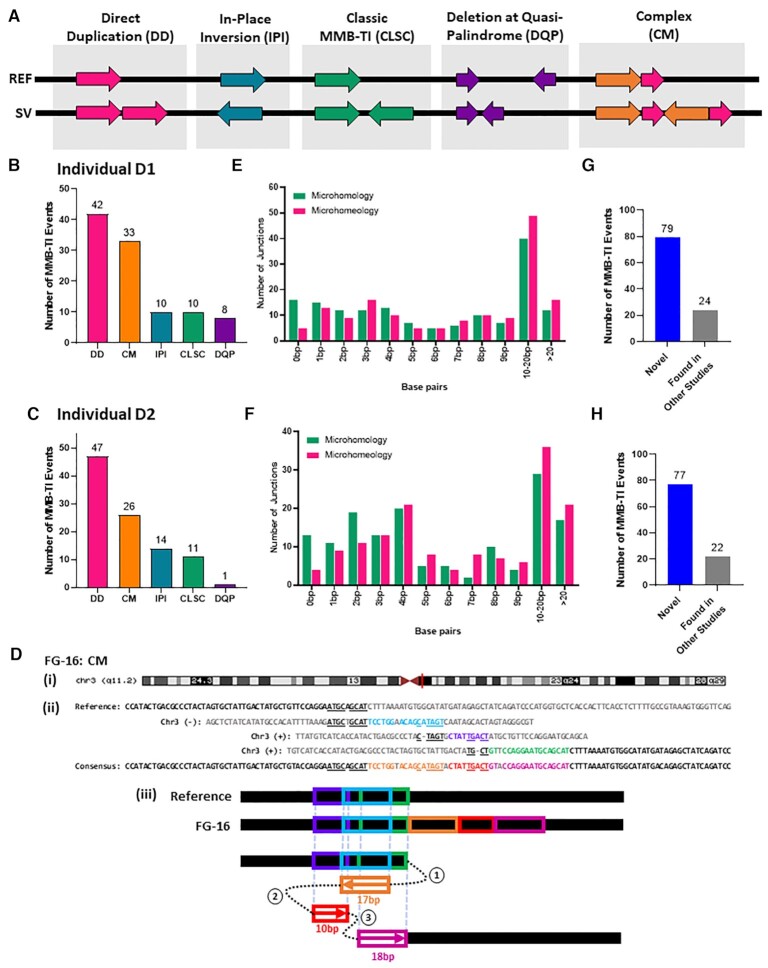
Figure 2.
MMB-TIs are found as germline variants in human fibroblasts. (A) Schematic of different classes of MMB-TI events. REF = reference; SV = MMB-TI event; (B) Distribution of different MMB-TI classes among germline events (parameters for search: ≥25 bp in length) identified in individual D1 (n = 103) (27). (C) Similar to (B), but for individual D2 (n = 99) (27). (D) (i) Complex MMB-TI (CM) event FG-16 found on q11.2 band of Chromosome 3 (red vertical line indicates position; maroon triangles indicate centromere location). (ii) Alignment of event FG-16 consensus to all refence templates. Insertion consists of three parts (orange, red and pink text) copied from all corresponding templates (light blue, purple and green respectively). Microhomology used in each template switching event is underlined. (iii) Schematic illustrating template switching events that lead to the formation of FG-16. Circled numbers indicate the order of template switches. Boxed arrows indicate direction of synthesis, numbers under boxes indicate length of synthesis, and colors correspond to those in (ii). (E) Distribution of microhomology analyzed for all resolvable junctions of MMB-TIs from D1 shown in (B) (n = 155). Microhomology = identical bases; microhomeology = identical, but with 1bp mismatch or gap allowed. (F) Similar to (E), but for D2 shown in (C) (n = 148). (G) Number of novel MMB-TIs from D1 (from B) and those already existing in NCBI’s Nucleotide collection (nr/nt). (H) Similar to (G), but for events from D2 (from C).