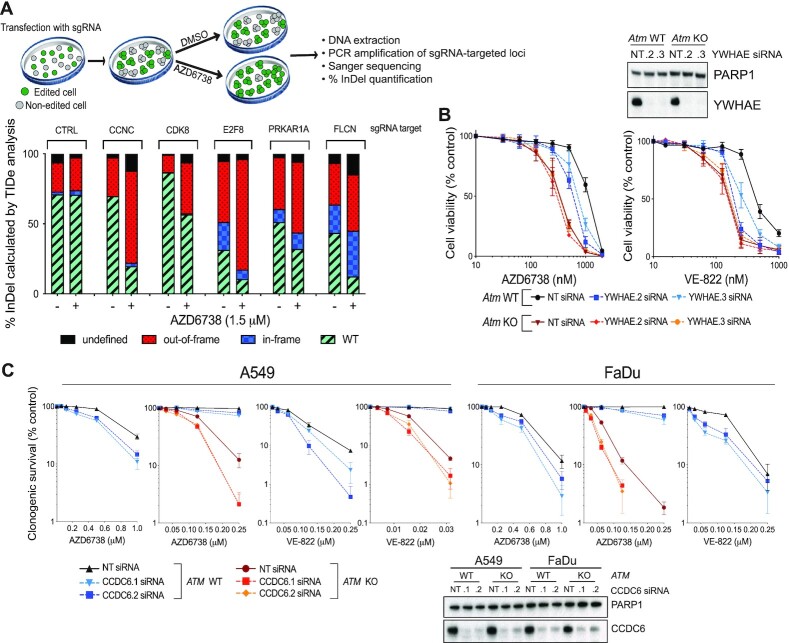
Figure 2.
Validation of hits identified through our CRISPR–Cas9 screens. (A) Competition assays between CRISPR–Cas9 edited and non-edited cells in response to AZD6738 treatment. Cas9-expressing mESCs were transfected with synthetic sgRNAs against the target loci, resulting in a mixed pool of edited and non-edited cells after 7 days. Each pooled population was treated for 5 days with DMSO or 1.5 μM AZD6738, with a high dose strongly enriching for resistant cells. Genomic DNA was extracted and the sgRNA-targeted loci PCR amplified to allow editing efficiencies and % InDels (–50 bp to +50 bp) to be calculated using TIDe software (3.2.0) (44). The control sgRNA targeted Lrrc29 which was not predicted to influence ATRi sensitivity in Atm WT mESCs according to our screening data. (B) Results of MTT cell proliferation assays in Atm WT and KO mESCs following siRNA-depletion of YWHAE, in response to two chemically distinct ATR inhibitors (AZD6738 and VE-822). Error bars = mean ± SEM (biological n = 3). (C) Clonogenic survivals of ATM WT and KO A549 and FaDu cells following siRNA-depletion of CCDC6, in response to two chemically distinct ATR inhibitors (AZD6738 and VE-822). Error bars = mean ± S.D (biological n = 3). FaDu ATM KO cells are inherently extremely sensitive to doses of VE-822 which effectively inhibit ATR kinase activity, therefore precluding analysis of further reductions in clonogenic survival upon CCDC6 depletion.