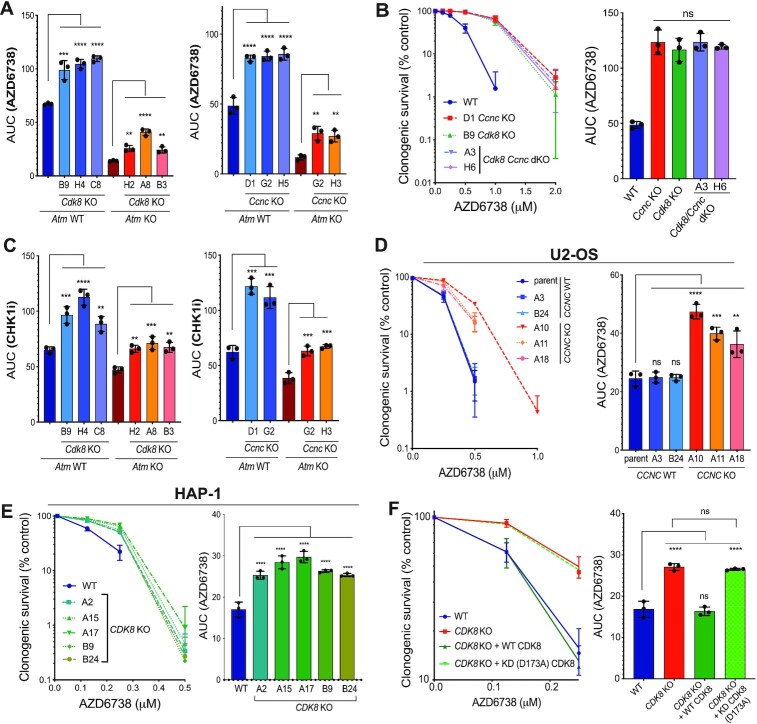
Figure 3.
Loss of Cyclin C/CDK8 provides ATRi and CHK1i resistance. (A) Clonogenic survivals of Ccnc/Cdk8 WT/KO Atm WT/KO mESCs treated with AZD6738, represented as AUCs (areas under the curve). Error bars = mean ± S.D (biological n = 3). Survival curves can be found in Supplementary Figure S3D. (B) Clonogenic survivals of WT and Ccnc/Cdk8 single and dual KO mESCs treated with AZD6738. Data also represented as AUCs. Error bars = mean ± SD (biological n = 3). (C) Clonogenic survivals of Ccnc/Cdk8 WT/KO Atm WT/KO mESCs treated with the CHK1 inhibitor LY2603618, represented as AUCs. Error bars = mean ± SD (biological n = 3). Survival curves can be found in Supplementary Figure S3F. (D) Clonogenic survivals of WT and CCNC KO U2-OS cells in response to AZD6738 treatment. A3/B24 clones were confirmed as WT by immunoblot analyses and genotyping after single-cell expansions of sgRNA-transfected cells. Data also represented as AUCs. Error bars = mean ± SD (biological n = 3). (E) Clonogenic survivals of WT and CDK8 KO HAP-1 cells in response to AZD6738 treatment. Data also represented as AUCs. Error bars = mean ± SD (biological n = 3). (F) Clonogenic survivals of WT and CDK8 KO HAP-1 cells, complemented with WT or kinase dead (D173A) CDK8-GFP, treated with AZD6738. Data also represented as AUCs. Error bars = mean ± SD (biological n = 3). Additional clones are shown in Supplementary Figure S5A and B. All statistical analyses were performed using a one-way ANOVA test with multiple comparisons. P-values < 0.05 (*), 0.01 (**), 0.001 (***) and 0.0001 (****) were deemed statistically significant.