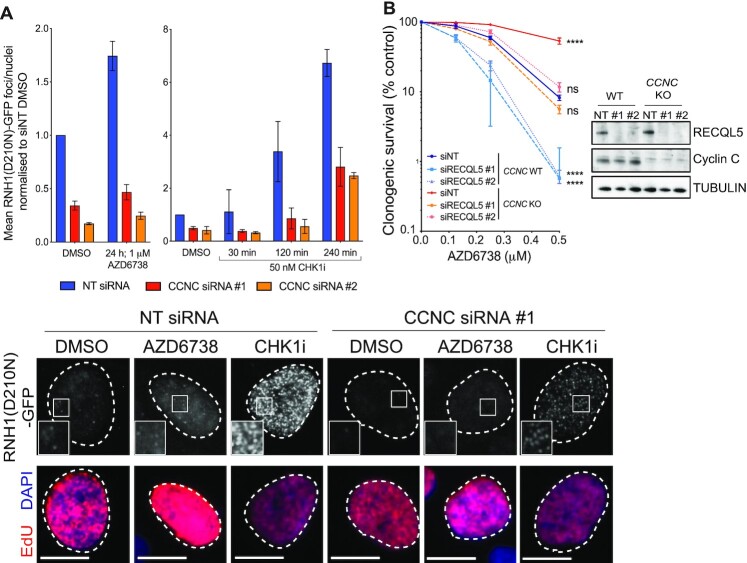
Figure 5.
Cyclin C loss supresses DNA:RNA hybrid formation in response to ATRi and CHK1i. (A) Mean number of RNH1(D210N)-GFP foci/nuclei normalised to untreated NT siRNA levels in response to AZD6738 or CHK1i (LY2606368) treatment, following siRNA-depletion of Cyclin C. Error bars = mean ± SD (biological n = 2). Representative images are of RNH1(D210N)-GFP foci in EdU positive cells, following treatment with DMSO, AZD6738 for 24 h, or CHK1i for 4 h. Scale bars = 12 μm. A full field of view is provided in Supplementary Figure S7C which also highlights the heterogeneity of foci induction in Cyclin C-depleted cells treated with CHK1i. SiRNAs targeting CDK8 were also tested but resulted in the accumulation of cells in G1 which may bias the experimental outcome (Supplementary Figure S7A and B). (B) Clonogenic survivals of RECQL5-depleted WT and CCNC KO U2-OS cells treated with AZD6738. Error bars = mean ± SD (biological n = 3). Statistical analyses were performed using a two-way ANOVA test by comparison to CCNC WT cells transfected with the control (NT) siRNA. P-values < 0.05 (*), 0.01 (**), 0.001 (***) and 0.0001 (****) were deemed statistically significant.