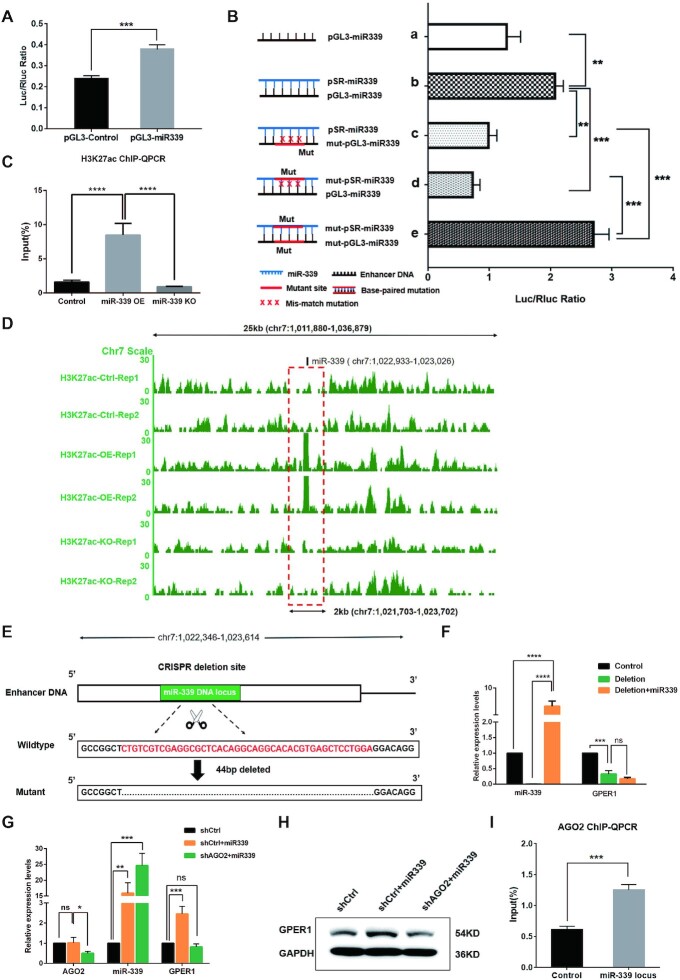
Figure 4.
MiR-339 reactivates TSG GPER1 depended on Enhancer elements and AGO2 protein. (A, B) MiR-339 genome locus acts as an enhancer by Dual Luciferase Reporter Assay. (A) The activity of reporter gene was activated by pGL3-miR339 in HEK293T cells. (B, a, b) The ratio of Luc+/Rluc + was remarkably higher for the experimental group with co-transfecting pGL3-miR339 and pSR-miR339 than that of the control group with co-transfecting pGL3-miR339 and pSUPER empty plasmids; (b–d) The luciferase activity was reduced by either mutating enhancer elements (mut-pGL3-miR339) or mutating seed sequence of miR-339 (mut-pSR-miR339); (e) The luciferase activity was rescued by mutating both the enhancer and seed sequence of miR-339 with a matched complementary base pair. (C, D) The enrichment of H3K27ac in enhancer region containing miR-339 DNA locus by ChIP-qPCR assay (C) and H3K27ac ChIP-seq analyses (D). (E, F) The activation was abolished by deletion of the miR-339 enhancer region in HEK293T cells by CRISPR/Cas9. schematic diagram (E) shows a deletion of 44bp on enhancer region by CRISPR/Cas9 system, while miR-339 (F) failed to reactivate GPER1 expression. (G, H) AGO2 is essential to the activating function of miR-339 on GPER1 expression. GPER1 was no longer activated by miR-339 in RT-PCR (G) and western blot assays (H) by knocking down AGO2 in HEK293T cells. (I) AGO2 was recruited to miR-339 locus when transfected miR-339 by ChIP-qPCR assay. Data are presented as Mean ± SD of experiments conducted in triplicate. (C, F, G) One-ANOVA test, followed by Tukey's test. ****P <0.0001, ***P <0.001, **P <0.01, *P <0.05. “ns” is representative of no significance.