Aire is a master of central tolerance. Goldfarb et al. compare mice with recessive or dominant-negative Aire patient mutations to dissect the mechanisms that underlie dominance and identify an autoregulatory mechanism by which Aire negatively modulates its own expression.
Abstract
The autoimmune regulator (AIRE) is essential for the establishment of central tolerance and prevention of autoimmunity. Interestingly, different AIRE mutations cause autoimmunity in either recessive or dominant-negative manners. Using engineered mouse models, we establish that some monoallelic mutants, including C311Y and C446G, cause breakdown of central tolerance. By using RNAseq, ATACseq, ChIPseq, and protein analyses, we dissect the underlying mechanisms for their dominancy. Specifically, we show that recessive mutations result in a lack of AIRE protein expression, while the dominant mutations in both PHD domains augment the expression of dysfunctional AIRE with altered capacity to bind chromatin and induce gene expression. Finally, we demonstrate that enhanced AIRE expression is partially due to increased chromatin accessibility of the AIRE proximal enhancer, which serves as a docking site for AIRE binding. Therefore, our data not only elucidate why some AIRE mutations are recessive while others dominant, but also identify an autoregulatory mechanism by which AIRE negatively modulates its own expression.
Introduction
The autoimmune regulator (AIRE) is a unique transcription regulator, essential for the induction of self-tolerant T cells during their development in the thymus (Anderson et al., 2002; Mathis and Benoist, 2009). AIRE is expressed almost exclusively in mature medullary thymic epithelial cells (mTECs), where it promotes expression of hundreds of different tissue-restricted antigen (TRA) genes for the purpose of negatively selecting autoreactive thymocytes or diverting them to a T regulatory lineage (Anderson et al., 2002; Aschenbrenner et al., 2007; Mathis and Benoist, 2009; Savage et al., 2014). The importance of AIRE in these processes is best illustrated by the severe multi-organ autoimmune syndrome caused by its mutations (Finnish-German APECED Consortium, 1997; Nagamine et al., 1997). A similar multi-organ autoimmune phenotype can be induced by germline inactivation of Aire in mice and rats, although the repertoire of targeted tissues differs between species and/or genetic backgrounds (Anderson et al., 2002; Jiang et al., 2005; Ossart et al., 2018).
Historically, the disease caused by AIRE mutations named autoimmune polyendocrine syndrome type 1 (APS-1; OMIM: 240300) was considered to follow an autosomal-recessive mode of inheritance. However, in 2001, an Italian cohort was described with a dominant inheritance pattern that segregated with a monoallelic G228W missense mutation (Cetani et al., 2001) in AIRE’s putative DNA-binding domain named SAND after proteins containing such a domain (Sp100, AIRE, NucP41/75, and Deaf1; Gibson et al., 1998). This suggested that some AIRE mutations may operate in a dominant-negative manner with incomplete penetrance (Cetani et al., 2001). Indeed, subsequent characterization of this mutation in a mouse model (Aire+/G228W) confirmed this hypothesis (Su et al., 2008). More recently, AIRE’s first plant homeodomain (PHD1), which has histone-reading capacity (Koh et al., 2008; Org et al., 2008), was found to be a hotspot for multiple dominant-negative mutations, including V301M, C302Y, G305S, C311Y, R316W, P326L, and R328Q (Oftedal et al., 2015). In general, individuals with monoallelic PHD1 and SAND mutations present with milder autoimmune manifestations than classical APS-1 (Abbott et al., 2018; Cetani et al., 2001; Oftedal et al., 2015), with a preponderance for pernicious anemia, vitiligo, and type 1 diabetes, but the mechanisms behind the dominant-negative effects are unknown.
Therefore, to provide further mechanistic insights, we used CRISPR/Cas9 genome editing and generated several novel mouse models, each bearing either a known recessive or putative dominant-negative AIRE patient mutation. Indeed, we have experimentally established that some monoallelic AIRE mutants, including Aire+/C313Y and Aire+/C442G, cause breakdown of central tolerance and give rise to multiorgan autoimmunity. Moreover, by using various genomic and protein analyses, we dissect the underlying mechanisms for their dominancy. Specifically, we show that while the recessive mutations result in a lack of AIRE expression, the dominant mutations invariably augment the expression of dysfunctional AIRE with effects on subcellular localization, capacity to bind chromatin, and induction of gene expression determined by the specific dominant-negative mutation. Furthermore, these data reveal unique contributions of the PHD1 versus PHD2 domains of AIRE in transcriptional regulation. Finally, we also identify a novel autoregulatory mechanism by which AIRE negatively modulates its own expression.
Results
Heterozygous Aire+/C313Y mutation affects fertility and mTEC cellularity
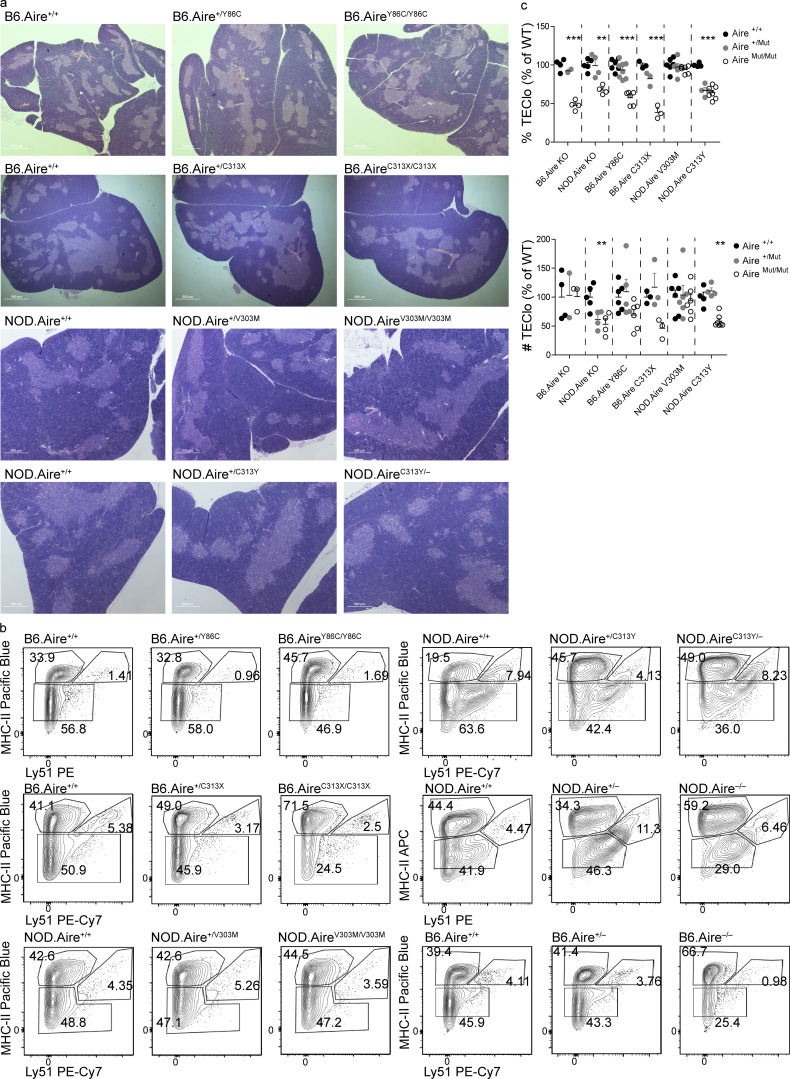
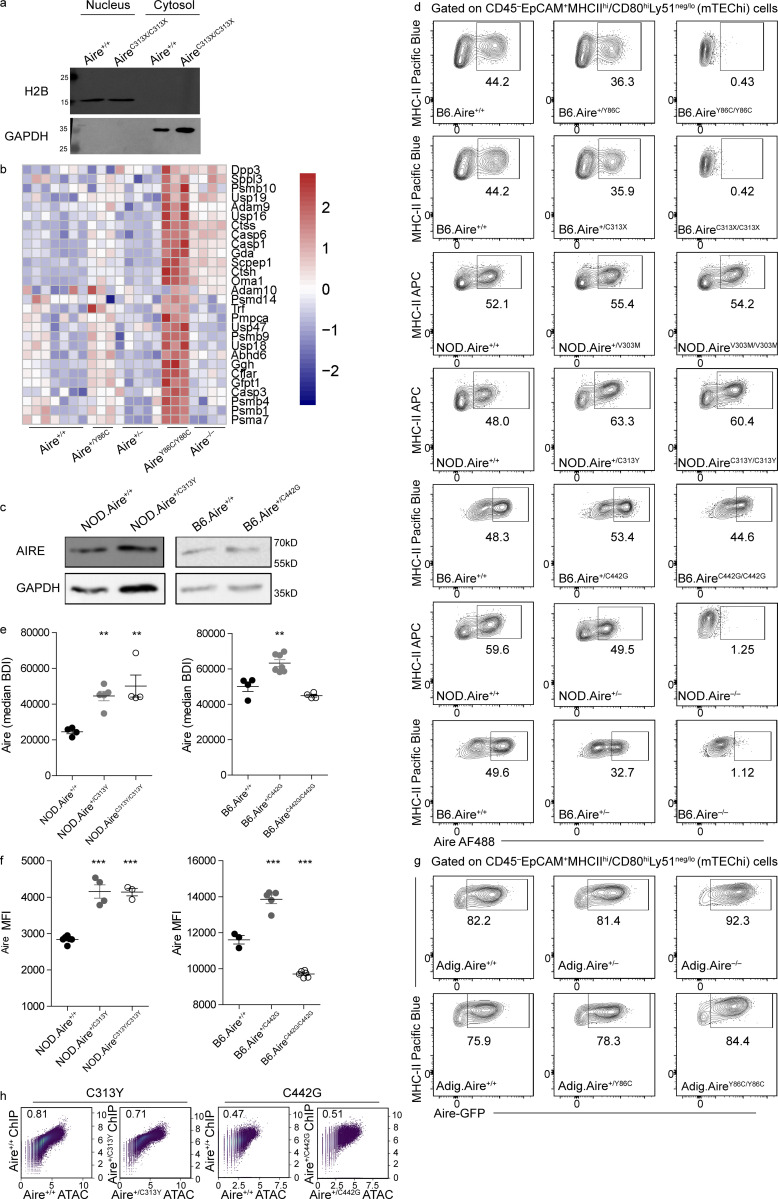
Although several dominant mutations in AIRE PHD1 have been reported in patients and families with diverse autoimmune manifestations (Oftedal et al., 2015), our understanding of the molecular mechanisms underlying their dominancy remains largely elusive. Therefore, in order to assess the specific effects of the putative dominant-negative mutations, we generated two mouse models corresponding to the V301M and C311Y AIRE PHD1 human mutations (V303M and C313Y, respectively, in the mouse) on the nonobese diabetic (NOD) background using CRISPR/Cas9 genome editing (Fig. 1 a and Table S1). The autoimmune-prone NOD background was selected to allow more comprehensive assessment of prospective autoimmunity caused by each mutation. The V301M mutation was chosen due to its relatively high prevalence among the general population (0.00089; Oftedal et al., 2015), while the C311Y mutation was chosen due to its robust patient phenotype compared with the other PHD1-dominant mutations (Oftedal et al., 2015). Importantly, previous solution of the AIRE PHD1 structure showed that only the C311Y mutation, but not V301M, disrupts the PHD1 fold, as the original cysteine is critical for binding one of the Zn2+ ions at the center of the domain and maintaining proper fold, while its substitution for tyrosine interrupts Zn2+ binding, thus impairing domain function (Bottomley et al., 2005; Gaetani et al., 2012; Koh et al., 2008; Fig. 1 b). Furthermore, to understand the differences between the dominant-negative and recessive mutations, we generated mouse models of two known recessive mutations, Y85C (Y86C in mice) and C311X mutation (C313X in mice; Björses et al., 2000; Oftedal et al., 2015; Fig. 1, a and b). Y85C is located in the caspase recruitment domain (CARD; Ferguson et al., 2008), essential for AIRE’s ability to form homo-dimers or homo-tetramers (Björses et al., 2000; Oftedal et al., 2015). The C311X mutation, which has a premature termination codon (PTC) in the PHD1 domain, was of particular interest, as it affects the same cysteine as C311Y, allowing us to better address and understand why two different mutations of the codon subsequently exert recessive or dominant capacities.
Figure 1.
Heterozygous Aire+/C311Y mutation affects fertility and mTEC cellularity. (a) Scheme of AIRE protein domains (top) with human point mutations highlighted and the respective mouse mutants created in parentheses. Chromatograms of DNA sequences around the point mutations created in AIRE mutant mice. The relevant patient-based nucleotide changes between the WT (middle) and homozygous (bottom) sequences are highlighted by black triangles. Gray triangles indicate silent mutations inserted by the repair oligo to mutate protospacer adjacent motif sequences. (b) Models of human WT (top) and mutated (bottom) AIRE domain structures with mutated amino acids in red and the WT amino acids in green. As the CARD domain structure has yet to be solved, the CARD structure was estimated based on comparison to other proteins with CARD domains. Within the CARD structure, key hydrophobic residues (Ferguson et al., 2008) are in orange, and LXXLL motifs are pink. Within the PHD1 (Bottomley et al., 2005), Zn2+ ions are teal spheres, with the supporting histidines and cysteines in yellow. The disrupted Zn2+ ligation of the C311Y mutation is highlighted by a black arrowhead. (c) Average number of pups born to NOD.Aire+/− or NOD.Aire+/C313Y females mated with NOD.Aire+/C313Y males over the course of ∼2 mo. Data are from 7–11 mice per group, analyzed by Student’s t test, and are represented as mean ± SEM, ***, P < 0.001. (d and e) Frequencies (d) and absolute counts (e) of mTEChi (EpCAM+CD45–MHCIIhiLy51–/lo) in the different AIRE mutants. Data from are from two to eight mice per group, analyzed by one-way ANOVA, and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from the relevant WT littermate control. Both frequencies and absolute counts are calculated as a percentage from the average frequency or count of all WT animals within a given experiment. Each mouse strain was evaluated separately. At least two independent experiments were conducted on each strain.
All strains were then bred in Aire+/Mut × Aire+/Mut settings and gave progeny with normal frequencies and Mendelian ratios, except for the Aire+/C313Y strain. Specifically, in a comparison between NOD.Aire+/C313Y and NOD.Aire+/− females bred with NOD.Aire+/C313Y males, a significant reduction in the number of pups born over a period of ∼2 mo was seen for the NOD.Aire+/C313Y females, which also did not produce more than one litter, if at all (Fig. 1 c). Therefore, in order to maintain this strain, NOD.Aire+/C313Y males were bred with NOD.Aire+/− or NOD.Aire+/+ females. Since we were unable to produce sufficient numbers of AireC313Y/C313Y animals for all experiments, we used AireC313Y/– mice for the assessment of the impact of C313Y alone (i.e., without the presence of the WT allele) in most subsequent experiments. The almost complete failure to breed and to produce progeny of NOD.Aire+/C313Y females corresponds well with breeding problems described for Aire−/− animals (Jasti et al., 2012), and supports a dominant-negative effect of the C311Y mutation.
To further characterize the individual strains, we next examined their gross thymic morphology, which was normal in all mutant strains, as seen by the presence and clear demarcation between cortex and medulla (Fig. S1 a). However, more detailed analysis by flow cytometry highlighted different effects of the individual monoallelic mutations on the composition of the thymic epithelial compartment. As expected, we observed no differences in absolute numbers and frequencies of mature mTECs (mTEChi, EpCAM+CD45–MHCIIhiLy51–/lo) between Aire+/+ and their Aire+/Y86C or Aire+/C313X heterozygous littermates. Conversely, their homozygous AireY86C/Y86C or AireC313X/C313X counterparts revealed a significant increase in both the frequency and absolute counts of mTEChi (Fig. 1, d and e; and Fig. S1 b), similar to that seen in Aire−/−. More importantly, while no differences were noted in NOD.Aire+/V303M or NOD.AireV303M/V303M mice compared with their NOD.Aire+/+ counterparts (Fig. 1, d and e), a significant increase in both absolute number and frequency of mTEChi, and a concomitant reduction in the frequency of TECs expressing low levels of MHCII (TEClo, EpCAM+CD45–MHCIIlo/midLy51–/lo), were noted in both NOD.Aire+/C313Y and NOD.AireC313Y/– (Fig. 1, d and e; and Fig. S1, b and c), similar to earlier reports on Aire−/− mice (Hubert et al., 2009; St-Pierre et al., 2015; Fig. 1, d and e; and Fig. S1, b and c). Taken together, these data show that the thymic morphology of Aire+/C313Y is similar to Aire−/− and suggest that Aire+/C313Y is indeed dominant, while questioning the dominancy of the common Aire+/V303M mutation.
Figure S1.
Heterozygous Aire+/C311Y mutation affects fertility and mTEC cellularity. (a) Representative H&E staining of thymi from young mice (4–6 wk old) from the Aire Y86C, C313X, V303M, and C313Y mouse strains created taken at 2× magnification. (b) Representative flow cytometry plots of the TEC compartment of Aire Y86C, C313X, V303M, and C313Y mouse strains created, in addition to Aire−/− on both B6 and NOD backgrounds. Numbers indicate the percentage of cells within each gate from the entire TEC compartment. (c) Frequencies (top) and absolute counts (bottom) of TEClo (EpCAM+CD45–MHCIIlo/midLy51–/lo) of Aire Y86C, C313X, V303M, and C313Y mouse strains created, in addition to Aire−/− on both B6 and NOD backgrounds. Data are from two to eight mice per group, analyzed by one-way ANOVA and represented as mean ± SEM. **, P < 0.01; ***, P < 0.001 from the relevant WT littermate control. Both frequencies and absolute counts are calculated as a percentage from the average frequency or count of all WT animals within a given experiment. Each mouse strain was evaluated separately. At least two independent experiments were conducted on each strain.
Heterozygous Aire+/C313Y mutation, but not Aire+/V303M, impairs ectopic expression of AIRE target genes in mTECs and the thymic regulatory T (T reg) cell compartment
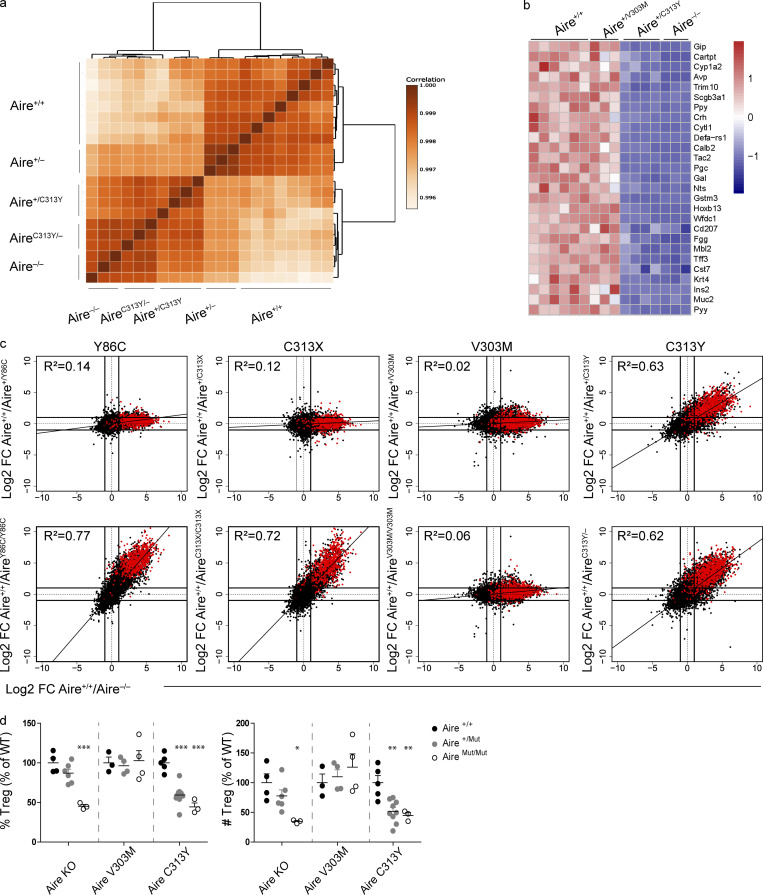
To further examine the dominant-negative effect of the AIRE PHD1 mutants, bulk RNA sequencing (RNAseq) of sorted mTEChi from the individual mutant strains or from their Aire−/−, Aire+/−, and Aire+/+ counterparts was performed. Interestingly, while no distinct clustering was found to segregate total gene expression between Aire+/+, Aire+/V303M, or AireV303M/V303M (Fig. S2 a), the Aire+/C313Y mTEChi uniquely clustered with AireC313Y/– mice harboring one allele with the C313Y mutation along with a null allele, thus mimicking C313Y homozygous mice, and Aire−/− mTEChi, while those from Aire+/− clustered with Aire+/+ (Fig. 2 a). Moreover, comparison of expression of representative Aire-dependent TRA genes validated a dramatic reduction in expression of Aire-dependent TRAs in Aire+/C313Y but not in the Aire+/V303M mTEChi (Fig. 2 b).
Figure S2.
Heterozygous Aire+/C313Y mutation, but not Aire+/V303M, impairs ectopic expression of AIRE target genes in mTECs and the thymic T reg cell compartment. (a) Dendrogram and heatmap of Pearson correlation coefficient between the different mTEChi (EpCAM+CD45−MHCIIhiLy51−/lo) bulk RNAseq samples based on the expression of all genes. NOD.Aire+/+, NOD.Aire+/V303M, and NOD.AireV303M/V303M (left), B6.Aire+/+, B6.Aire+/Y86C, and B6.AireY86C/Y86C (middle), and B6.Aire+/+, B6.Aire+/C313X, and B6.AireC313X/C313X (right). All mice assessed within a strain are littermates. (b) FC/FC plots of mTEChi gene expression comparing WT/heterozygous Aire mutants (left, y axis) to Aire+/+/Aire−/− (x axis) of the same background or WT/homozygous Aire mutants (right, y axis) to Aire+/+/Aire−/− (x axis) of the same background. All genes are depicted by black dots, while Aire-independent TRA genes are depicted by blue dots. (c) Heatmap of gene expression of 35 differentially expressed genes between NOD.Aire+/+ and NOD.AireV303M/V303M mTEChi. (d) Representative flow cytometry plots of thymocytes (top three panels) and T reg cells (CD4+CD8–CD25+Foxp3+; bottom three panels) from thymi of Aire V303M and C313Y mouse strains, in addition to NOD.Aire−/− mice. Numbers indicate the percentage of cells within the gate from the parent population. (e) Frequencies (left) and absolute counts (right) of CD4SP (CD4+CD8−), DP (CD4+CD8+), CD8SP (CD4−CD8+), and DN (CD4−CD8−) thymocyte populations. Data from three to nine mice per group, analyzed by one-way ANOVA, are represented as mean ± SEM. *, P < 0.05; **, P < 0.01 from the relevant WT littermate control. Both frequencies and absolute counts are calculated as a percentage from the average frequency or count of all WT animals within a given experiment. Each mouse strain was evaluated separately. NOD.AireC313Y/− mice were used in these experiments. At least two independent experiments were conducted on each strain.
Figure 2.
Heterozygous Aire+/C313Y mutation, but not Aire+/V303M, impairs ectopic expression of AIRE target genes in mTECs and the thymic T reg cell compartment. (a) Dendrogram and heatmap of Pearson correlation coefficient between the different mTEChi bulk RNAseq samples based on gene expression values. (b) Heatmap of normalized read counts of 27 representative AIRE-dependent genes in mTEChi of NOD.Aire+/+, NOD.Aire+/V303M, NOD.Aire+/C313Y, and NOD.Aire−/−. Expression is normalized per row. (c) FC/FC plots of mTEChi gene expression comparing WT/heterozygous AIRE mutants (top, y axis) to Aire+/+/Aire−/− (x axis) of the same background or WT/homozygous AIRE mutants (bottom, y axis) to Aire+/+/Aire−/− (x axis) of the same background. All genes are depicted by black dots along with a trendline for the best linear fit, and R2 is indicated in each plot. A signature of AIRE-dependent TRA genes, which are down-regulated by at least 10-fold in B6.Aire−/− (based on a dataset from Sansom et al., 2014) is superimposed using red dots. All animals within a strain are littermates. (d) Frequency (left) and absolute counts of T reg cells (right; CD4+CD8–CD25+Foxp3+) in thymi of different NOD AIRE mutants created in this study, as well as Aire−/− on a NOD background. Data from three to nine mice per group, analyzed by one-way ANOVA, are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from the relevant WT littermate control. Both frequencies and absolute counts are calculated as a percentage from the average frequency or count of all WT animals within a given experiment. Each mouse strain was evaluated separately. At least two independent experiments were conducted on each strain.
To get a more comprehensive picture of the impact of the individual mutations on AIRE-mediated gene expression in mTECs, we performed fold change–fold change (FC/FC) analysis of the mTEChi RNASeq datasets obtained from the individual AIRE mutants (Fig. 2 c). Indeed, this analysis highlighted a clear diagonal pattern in the FC/FC plot between Aire+/+ versus Aire+/C313Y in comparison to Aire+/+ versus Aire−/− (Fig. 2 c), demonstrating that the impact of Aire+/C313Y on global gene expression largely overlaps with that of Aire−/−. The robust effect of Aire+/C313Y on gene expression and its similarity to that of AireC313Y/– are also seen by comparable correlations (R2) between gene expression profiles of Aire+/C313Y or AireC313Y/– with Aire−/− mTEChi (Fig. 2 c, top right and bottom right, respectively). This was further supported by superimposition of gene signatures of AIRE-dependent and AIRE-independent TRA genes based on a previously published RNAseq dataset (Sansom et al., 2014), which highlighted a clear shift in AIRE-dependent TRA genes (Fig. 2 c), but not in AIRE-independent TRA genes (Fig. S2 b). In accordance with the TEC cellular composition discussed above, no differences were seen in the expression of both AIRE-dependent and -independent TRAs between Aire+/+ and Aire+/V303M. Interestingly, even the homozygous AireV303M/V303M mutation had only a minor effect on the mTEChi transcriptome, with only 35 genes being differentially expressed compared with Aire+/+ (Fig. 2 c and Fig. S2 c), suggesting that AIREV301M represents a variant with relatively normal transcriptional activity. Finally, RNAseq of sorted mTEChi from the Y86C and C313X mice validated their recessive nature, as the impact of Aire+/Y86C or Aire+/C313X on gene expression was not different from that of Aire+/+ (Fig. 2 c; and Fig. S2, a and b), while AireY86C/Y86C or AireC313X/C313X was comparable to Aire−/− on the same background (Fig. 2 c; and Fig. S2, a and b). Collectively, these findings highlight the unique and robust dominant-negative effect of Aire+/C313Y on AIRE-driven promiscuous gene expression in the thymic epithelium.
Impaired promiscuous gene expression in Aire−/− mice was previously shown to subsequently result in an altered CD4+CD8–CD25+Foxp3+ T reg cell compartment (Aricha et al., 2011; Cowan et al., 2018; Yang et al., 2015). We therefore assessed the numbers and frequencies of T reg cells in thymi isolated from Aire+/C313Y and Aire+/V303M mutants. Indeed, we observed a significant reduction in both absolute numbers and frequency of T reg cells in Aire+/C313Y mice, similar to that observed in Aire−/− (Fig. 2 d and Fig. S2 d), while other thymocyte subpopulations remained largely unchanged (Fig. S2, d and e). In accordance with the lack of change in TRA gene expression in both Aire+/V303M and AireV303M/V303M mice, no differences were detected in the T reg cell absolute numbers or frequency in the thymi of these mice, further supporting that the V301M mutation is not dominant, in contrast to our previous prediction (Oftedal et al., 2015).
The dominant-negative Aire+/C313Y mutation results in devastating multi-organ autoimmunity
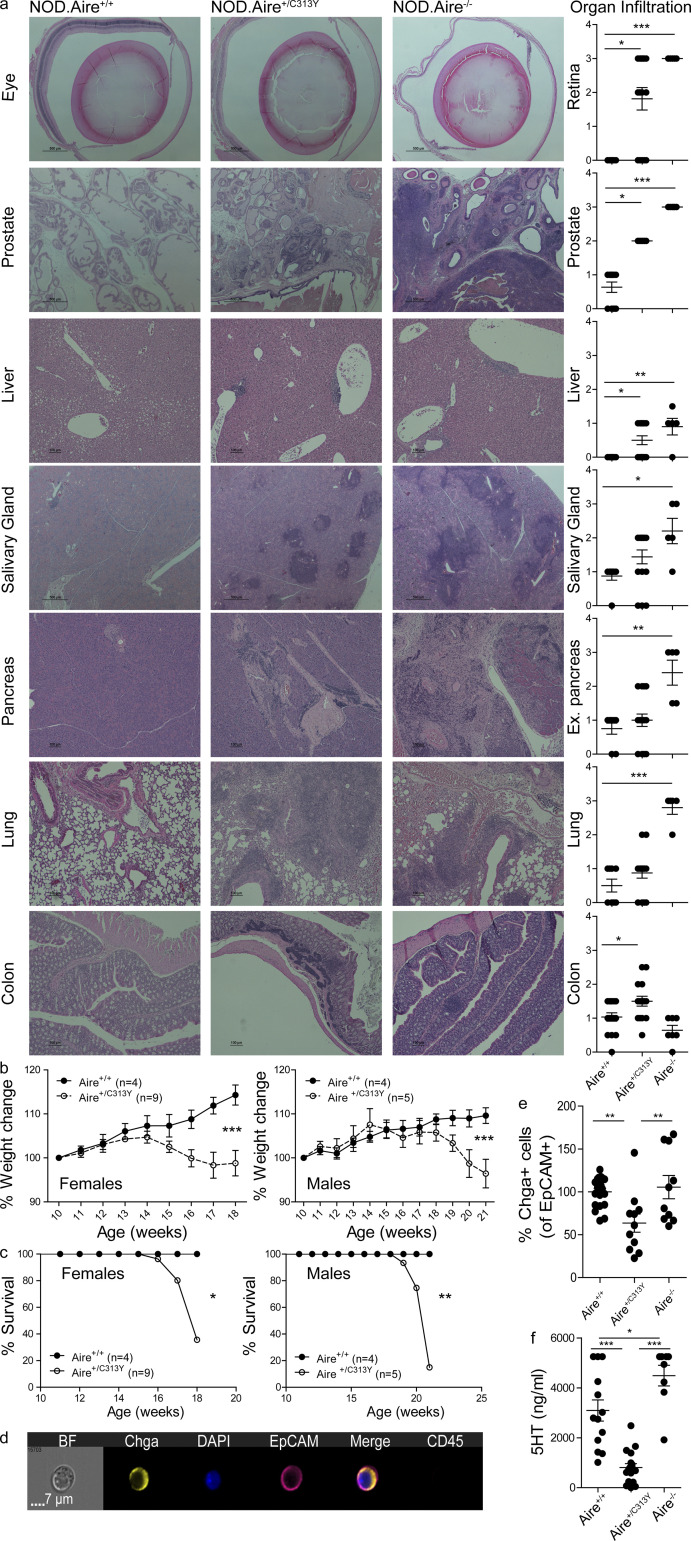
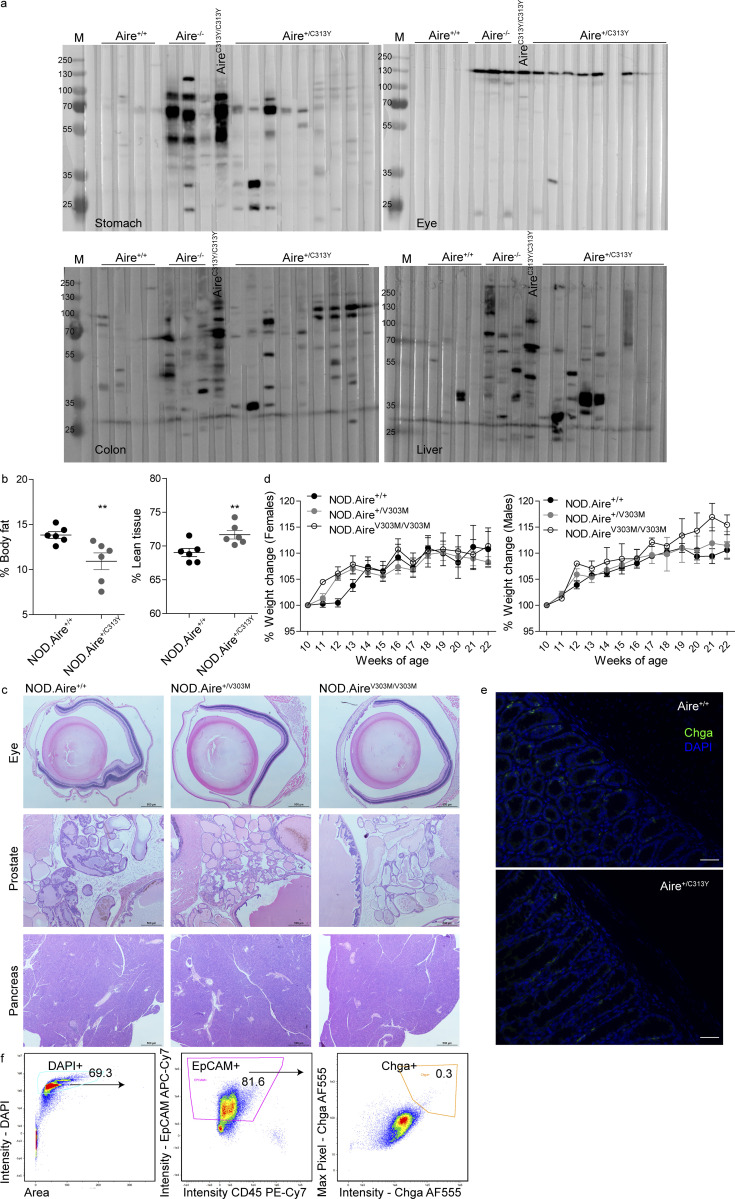
To validate whether the significant reduction in AIRE-dependent gene expression and T reg cells in the thymi of Aire+/C313Y mice translated into organ-specific autoimmunity, we aged the corresponding mice for 14–21 wk and analyzed autoimmunity against multiple tissues. Indeed, lymphocytic infiltrates were readily detected in the liver, prostate, and salivary glands of aged Aire+/C313Y mice, as was severe retinal degeneration (Fig. 3 a). Aged Aire+/C313Y mice also developed autoantibodies against various peripheral organs such as the eye, stomach, and liver (Fig. S3 a). While the organs affected in Aire+/C313Y mice largely mirrored those targeted in Aire−/− mice, the degree of infiltration and tissue destruction were overall milder, as seen by reduced characteristic infiltration of the exocrine pancreas and the lungs, to which the wasting of NOD.Aire−/− mice is attributed (Jiang et al., 2005). Nevertheless, the autoimmune attack in Aire+/C313Y mice resulted in fatal wasting disease, as demonstrated by total weight loss over time, with females wasting before males (Fig. 3 b). Specifically, we observed a reduction in the percentage of body fat in Aire+/C313Y 21-wk-old males with concomitant elevation in the percentage of lean tissues (including internal organs, muscle, bone, and connective tissues; Fig. S3 b). The deterioration culminated in reduced survival rates in Aire+/C313Y mice of both sexes compared with their Aire+/+ littermates (Fig. 3 c). As expected from the mTEChi transcriptome analysis, no autoimmunity or wasting was detected in the Aire+/V303M mice (Fig. S3, c and d).
Figure 3.
The dominant-negative Aire+/C313Y mutation results in devastating multi-organ autoimmunity. (a) Representative H&E staining of histopathology in target organs from aged (16–21-wk-old) NOD.Aire+/+, NOD.Aire+/C313Y, and NOD.Aire−/− mice, as well as scoring of all mice per organ. Images were captured at 20× (colon), 10× (liver, pancreas, prostate, and lung), and 4× (eye and salivary gland) magnification. Data from 5–16 mice per group were analyzed by Kruskal–Wallis nonparametric test with Dunn’s post hoc test and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Ex., exocrine. (b) Weight change curves of NOD.Aire+/C313Y and NOD.Aire+/+ female (left) and male (right) mice. Weight change was calculated per mouse from its weight at 10 wk of age. Data from four to nine mice per group were analyzed by repeated-measures ANOVA and are represented as mean ± SEM at each time point. ***, P < 0.001 from WT littermate controls. (c) Survival curves of NOD.Aire+/C313Y and NOD.Aire+/+ female (left) and male (right) mice. Data from four to nine mice per group were analyzed by log-rank (Mantel–Cox) test. *, P < 0.05; **, P < 0.01 from WT littermate controls. (d) Representative snapshot of a Chga+ enteroendocrine cell analyzed by imaging flow cytometry. DAPI was added following fixation to allow nucleus visualization. BF, bright field. (e) Frequency of colonic Chga+ enteroendocrine cells of aged (16–21 wk) NOD.Aire+/+, NOD.Aire+/C313Y, and NOD.Aire−/− mice measured by imaging flow cytometry. Frequencies are calculated as a percentage of the average frequency of all WT animals within a given experiment. Data from three combined experiments with a total of 10–20 mice per group, analyzed by one-way ANOVA, are represented as mean ± SEM. **, P < 0.01. WT animals are littermates of either NOD.Aire+/C313Y or NOD.Aire−/− mice. (f) Levels of serum 5HT in aged (16–21 wk) NOD.Aire+/+, NOD.Aire+/C313Y, and NOD.Aire−/− mice measured by ELISA. Data from 8–17 mice per group were analyzed by one-way ANOVA and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S3.
The dominant-negative Aire+/C313Y mutation results in devastating multi-organ autoimmunity. (a) Slotblots of stomach (top left), eye (top right), colon (bottom left), and liver (bottom right), probed with serum from aged (14–21 wk) NOD.Aire+/+, NOD.Aire−/−, NOD.AireC313Y/C313Y, and NOD.Aire+/C313Y mice. M, marker. (b) Percentage of body fat (left) and lean tissues (including internal organs, muscle, bone, and connective tissues; left) measured in aged (21-wk-old) male NOD.Aire+/+ or NOD.Aire+/C313Y mice. Data from six mice per group, analyzed by Student’s t test, are represented as mean ± SEM. **, P < 0.01 marks significant differences from the relevant WT littermate controls. (c) Representative H&E staining of target organs (eye, prostate, pancreas, top to bottom) from aged (22-wk-old) NOD.Aire+/+, NOD.Aire+/V303M, and NOD.AireV303M/V303M mice. Images were captured at 4× magnification. Scale bars, 500 μm. (d) Weight change curves of NOD.Aire+/+, NOD.Aire+/V303M, and NOD.AireV303M/V303M female (left) and male (right) mice from the age of 10 wk until the end of experimentation. Weight change was calculated per mouse from its weight at 10 wk of age. Data from two to eight mice per group were analyzed by repeated-measures ANOVA and are represented as mean ± SEM at each time point. (e) Immunofluorescence staining of colon parafilm-embedded sections from aged (17-wk-old) NOD.Aire+/+ and NOD.Aire+/C313Y mice. Enteroendocrine cells stain positive for Chga (green). Cell nuclei were counterstained with DAPI. Images were captured at 20× magnification. Scale bars, 50 μm. (f) Gating strategy of colonic enteroendocrine cells using ImageStream. Numbers within gates represent the percentage from parent population. DAPI was added after fixation to allow nuclei visualization. Maximum pixel is defined as the highest intensity pixel within the image.
Wasting of the Aire+/C313Y mice at around 4–5 mo of age without significant destruction of the exocrine pancreas or pulmonary infiltration was striking and surprising, and led us to further examine the intestinal phenotype of these mice. Previous studies have shown autoimmune attack on enteric α-defensins and loss of Paneth cells in both APS-1 patients and Aire−/− mice (Dobeš et al., 2015), and targeting of enteroendocrine cells in APS-1 patients with gastric symptoms (Ekwall et al., 1998; Posovszky et al., 2012). As colons of a cohort of aged NOD.Aire+/C313Y mice showed significant lymphocytic infiltration (Fig. 3 a) coupled with marked production of colon-specific autoantibodies (Fig. S3 a), we examined the enteroendocrine compartment of the colon in aged NOD.Aire+/C313Y mice. To this end, we first examined frozen colon sections of aged Aire+/C313Y mice and their WT littermates for the presence of chromogranin A (Chga), which is unvaryingly expressed in all enteroendocrine cells (Fig. S3 e). At first glance, Chga+ cells in Aire+/C313Y mice seemed to be reduced in frequency compared with controls (Fig. S3 e). To further validate this observation, we employed imaging flow cytometry and found a significant reduction in the frequency of these cells (Fig. 3 d) in the colons of aged Aire+/C313Y but not in Aire+/+ or Aire−/− mice (Fig. 3 e). As >70% of enteroendocrine cells in the colon have been reported to express serotonin (5-hydroxytryptamine [5HT]), and gastrointestinal enteroendocrine cells account for >90% of peripheral 5HT production in the body with impacts on energy homeostasis and tissue repair (Gunawardene et al., 2011; Mann and Oakley, 2013; Moran et al., 2008), we measured and found significantly reduced levels of peripheral 5HT, indicating specific damage of enteroendocrine cells in aged Aire+/C313Y mice, but not in Aire+/+ or Aire−/− mice (Fig. 3 f).
Collectively, these data validate the autoimmune phenotype of Aire+/C313Y mutants and highlight interesting differences between them and Aire−/− mice. While the Aire+/C313Y mice have milder autoimmune phenotypes in their exocrine pancreas and lungs, they seem to develop a more severe colonic phenotype, characterized by specific damage of the enteroendocrine compartment.
AIREC313Y has impaired subnuclear localization and chromatin binding
To further examine the underlying mechanisms that could explain the robust decrease in AIRE-dependent TRA gene expression and the breakdown of tolerance in mice carrying the monoallelic Aire+/C313Y mutation, we first examined colocalization of AIRE with promyelocytic leukemia (PML) bodies. These matrix-bound subnuclear structures were recently shown to associate with AIRE C311Y in transfected cells and bring about transcriptional repression of AIRE-dependent genes in these cells (Huoh et al., 2020). Indeed, imaging flow cytometry analysis showed that while WT AIRE does not colocalize with PML, significant colocalization of PML and AIRE was found in mTECs from Aire+/C313Y and even more from AireC313Y/C313Y mice (Fig. 4, a and b). These findings suggest that in mTECs, AIREC313Y has an altered subnuclear localization, as it preferentially sequesters into PML bodies. Such altered subnuclear localization may likely interfere with AIRE’s binding to chromatin and/or its transcription-transactivation potential.
Figure 4.
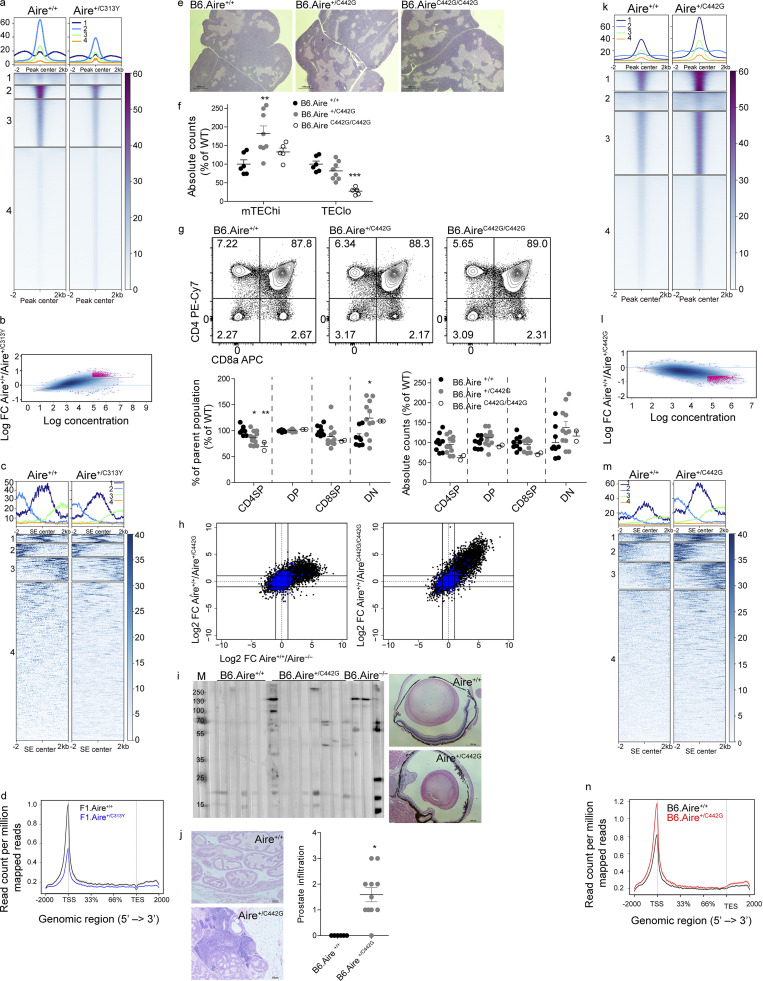
AIREC313Y has impaired subnuclear localization and chromatin binding. (a) Frequency of AIRE+ mTEChi with significant colocalization (i.e., bright detail similarity >1.5) of AIRE and PML in NOD.Aire+/+, NOD.Aire+/C313Y, and NOD.AireC313Y/C313Y littermate mice. Data from five to seven mice per group from two independent experiments, analyzed by one-way ANOVA, are represented as mean ± SEM. ***, P < 0.001 from the relevant WT littermate control. (b) Representative ImageStream snapshots of AIRE and PML colocalization in NOD.Aire+/+, NOD.Aire+/C313Y, and NOD.AireC313Y/C313Y littermate mice. BF, bright field. (c and d) Histogram for ChIPseq tag density for AIRE in F1.Aire+/+ and F1.Aire+/C313Y mTEChi superenhancer regions defined in Bansal et al. (2017) (c) and AIRE-dependent TRA genes (d; as in Fig. 2 c). (e) Normalized AIRE ChIPseq profiles of two representative superenhancer regions. The superenhancer boundaries are indicated by a red line. The range of normalized tag densities is indicated by the numbers in parentheses at the left of each track. To obtain enough mTEChi cells for ChIPseq, male NOD.Aire+/C313Y were bred with C57Bl/6 females to produce F1 progeny.
To gain insights into the association of dominant-negative AIREC313Y with chromatin, we performed AIRE chromatin immunoprecipitation followed by high-throughput sequencing (ChIPseq) in mTEChi isolated either from Aire+/+ or from Aire+/C313Y mice. Importantly, we observed ∼0.6 correlation between our data for WT AIRE and a previously published AIRE ChIPseq dataset using B6.Aire+/+ mTEChi (Bansal et al., 2017). Moreover, the technical validity of our data was further supported by a 0.77–0.88 Spearman correlation coefficient between the individual samples (Table S2). Strikingly, the analysis also revealed that while AIRE bound to 31,167 sites on average in the genome of Aire+/+ mTEChi, it bound a staggering average of 143,801 genomic sites in Aire+/C313Y mTEChi. Nevertheless, we observed lower read coverage of most Aire+/C313Y peaks compared with those of Aire+/+ (Fig. S4 a), supporting an important role for AIRE’s PHD1 domain in detection and/or binding to chromatin. Moreover, differential binding analysis of all peaks revealed lower read concentration and lower enrichment of binding sites for Aire+/C313Y compared with Aire+/+ (Fig. S4 b). It is well established that at a single-cell level, each mTEChi expresses only a fraction of the TRA repertoire, thereby creating a mosaic of heterogeneous cells. Consequently, Aire binding to predetermined superenhancer regions (Bansal et al., 2017), as well as TRA genes, is expected to be sparse within the bulk mTEChi population. Thus, our subsequent examination of these regions was based on read coverage density, rather than peak analysis. Interestingly, we observed decreased binding of the AIREC313Y mutant to superenhancer regions (Fig. 4, c and e). More detailed analysis of AIRE superenhancer binding also highlighted the differential intensities with which Aire+/+ and Aire+/C313Y bind different superenhancer regions (Fig. S4 c). Reduced binding of Aire+/C313Y was further mirrored by a clear decrease in the read occupancy at AIRE-dependent TRA genes in Aire+/C313Y compared with WT controls (Fig. 4 d). The effect was also apparent in AIRE-independent loci (Fig. S4 d), which were previously shown to have paradoxically higher occupancy of AIRE compared with its known TRA target genes (Bansal et al., 2017). Taken together, these findings suggest that in Aire+/C313Y mice, AIRE is incapable of efficiently binding both superenhancers as well as its target TRA genes and is rather preferentially localized to other genomic regions and PML bodies. Such altered chromatin localization is likely brought about by the misfolded histone-reading PHD1 domain, which impairs both AIRE binding to chromatin and subsequent AIRE-dependent gene expression and suggests that unlike WT AIRE, the C313Y dominant mutant is not likely to support looping between the enhancer sequences with their corresponding target genes.
Figure S4.
Identification of AIRE C446G as a novel dominant mutation located in the PHD2 domain. (a) Clustered heatmap of read coverage along all ChIPseq peaks in Aire+/+ and Aire+/C313Y samples. (b) MA plot comparing all ChIPseq binding regions of Aire+/+ and Aire+/C313Y, depicting the relationship between read density and FCs of binding regions. In pink are binding regions with false discovery rate <0.1. (c) Clustered heatmap of read coverage in mTEChi-specific superenhancer (SE) loci from Aire C313Y ChIPseq. (d) Histogram for ChIPseq tag density for AIRE in F1.Aire+/+ and F1.Aire+/C313Y mTEChi AIRE-independent TRA genes (as in Fig. S2 b). To obtain enough mTEChi cells for ChIPseq, male NOD.Aire+/C313Y were bred with C57Bl/6 females to produce F1 progeny. (e) Representative H&E staining of thymi from young (5-wk-old) B6.Aire+/+, B6.Aire+/C442G, and B6.AireC442G/C442G mice taken at 2× magnification. (f) Absolute counts of mTEChi (EpCAM+CD45–MHCIIhiLy51–/lo) and TEClo (EpCAM+CD45–MHCIIlo/midLy51–/lo) of B6.Aire+/+, B6.Aire+/C442G, and B6.AireC442G/C442G mice. Frequencies are presented as the percentage from the average frequency of all WT animals within a given experiment. Data from two combined experiments with a total of five to eight mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. **, P < 0.01; ***, P < 0.001 from the relevant WT littermate controls. (g) Representative flow cytometry plots of thymocytes (top) from B6.Aire+/+, B6.Aire+/C442G, and B6.AireC442G/C442G mice. Numbers indicate the percentage of cells within the gate from the parent population. Bottom panel shows frequencies (left) and absolute counts (right) of CD4SP (CD4+CD8–), DP (CD4+CD8+), CD8SP (CD4–CD8+), and DN (CD4–CD8–) thymocyte populations. Both frequencies and absolute counts are presented as a percentage from the average frequency or count of all WT animals within a given experiment. Data from two combined experiments with a total of 2–11 mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01 from the relevant WT littermate control. (h) FC/FC plots of mTEChi gene expression comparing B6.Aire+/+/B6.Aire+/C442G (left, y axis) to B6.Aire+/+/B6.Aire−/− (x axis), or B6.Aire+/+/B6.AireC442G/C442G (right, y axis) to B6.Aire+/+/B6.Aire−/− (x axis). All genes are depicted by black dots, while Aire-independent TRA genes are depicted by blue dots. (i) Slotblot of eye lysate probed with serum from aged (36–51-wk-old) B6.Aire+/+, B6.Aire+/C442G, and B6.Aire−/− mice (left) and representative H&E staining of histopathology in eyes from 36-wk-old B6.Aire+/+ and B6.Aire+/C442G mice (right, 4× magnification). M, marker. (j) Representative H&E staining of histopathology in prostate (10× magnification) from 36–51-wk-old B6.Aire+/+ and B6.Aire+/C442G mice (left), and as well as scoring. Data from 6–11 mice per group were analyzed by Kruskal–Wallis nonparametric test and are represented as mean ± SEM. *, P < 0.05 was considered statistically significant. (k) Clustered heatmap of read coverage along all ChIPseq peaks in Aire+/+ and Aire+/C442G samples. (l) MA plot comparing all ChIPseq binding regions of Aire+/+ and Aire+/C442G, depicting the relationship between read density and FCs of binding regions. In pink are binding regions with false discovery rate <0.1. (m) Clustered heatmap of read coverage in mTEChi-specific SE loci from Aire C442G ChIPseq. (n) Histogram for ChIPseq tag density for AIRE in B6.Aire+/+ and B6.Aire+/C442G mTEChi AIRE-independent TRA genes (as in h).
Identification of AIRE C446G as a novel dominant mutation located in the PHD2 domain
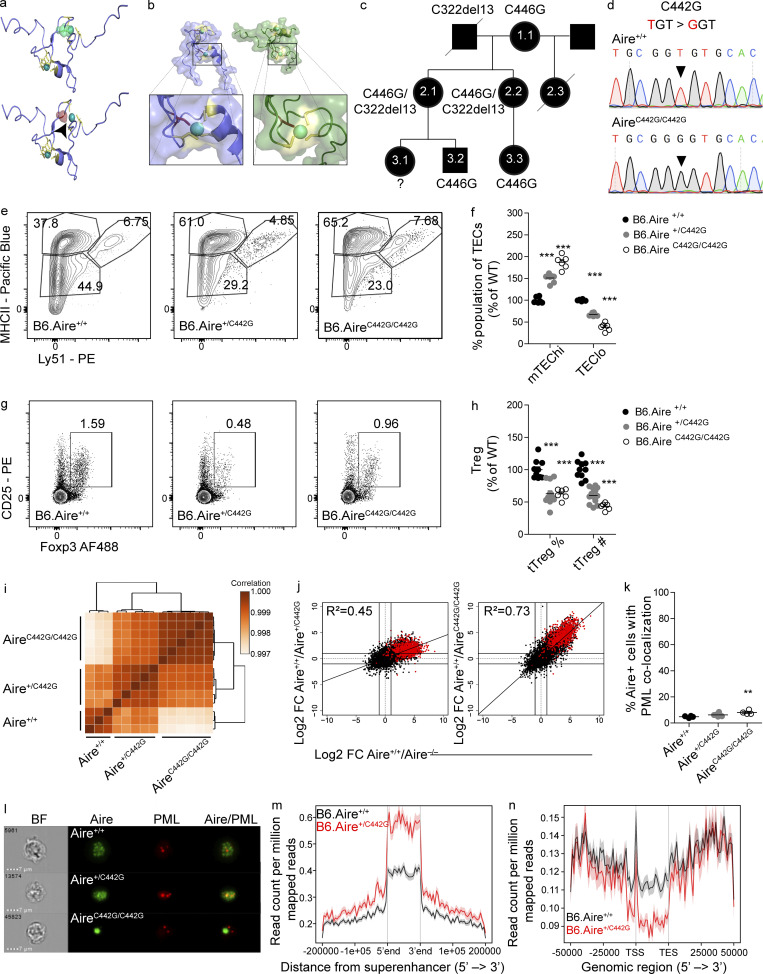
Validation of AIRE C311Y as a dominant-negative mutation prompted us to hypothesize that additional dominant mutations could be predicted based on their putative effect on AIRE structure. Nuclear magnetic resonance spectroscopy clearly demonstrated that AIRE C311Y distorts the fold of the PHD1 domain by preventing proper binding to one of the Zn2+ ions at the center of the domain (Bottomley et al., 2005). Another AIRE mutation, C446G (Wolff et al., 2007), was shown to have a similar effect by disrupting binding of an essential cysteine residue to the Zn2+ ion at the center of the PHD2 domain due to substitution for glycine, thus disrupting the conformation of AIRE’s PHD2 domain (Gaetani et al., 2012; Fig. 5 a). Given that PHD2 was previously shown to be indispensable for AIRE function (Yang et al., 2013), and that C446 is located in the analogous Zn2+-binding position in PHD2 as C311 is in PHD1 (Fig. 5 b), with partial dominant-negative activity in vitro (Oftedal et al., 2015), we hypothesized that the C446G mutation may exert a dominant-negative effect and reinvestigated APS-1 patients with C446G mutations. Indeed, in addition to the patient reported earlier with p.C446G/p.R257X diagnosed with classic APS-1 at 13 yr of age (Wolff et al., 2007), we identified another previously unreported patient (2.1) with p.C446G/p.C322del13 with APS-1 (Fig. 5 c and Table S3). Scrutiny of her family (Fig. 5 c) revealed a sister (2.2) with the same genotype harboring IFN-ω autoantibodies and severe inflammatory bowel disease (Table S3). Furthermore, the index patient’s mother (1.1) and a son (3.2) were monoallelic carriers of the C446G mutation. The mother had autoimmune thyroid disease, while the son at age 43 yr currently has no endocrinopathy. The daughter of the index person’s sister (3.3), a monoallelic carrier of C446G, has chronic diarrhea and Ehlers–Danlos disease. Interestingly, none of the individuals with monoallelic C446G had the IFN or organ-specific autoantibodies typically associated with autosomal-recessive APS-1 (Table S3; Oftedal et al., 2015). These data suggested that monoallelic AIRE C446G might also possess dominant-negative capacity, but with much milder autoimmune phenotypes.
Figure 5.
Identification of AIRE C446G as a novel dominant mutation located in the PHD2 domain. (a) The solution structure of the AIRE PHD2 domain (based on Gaetani et al., 2012), highlighting Zn2+-binding residues. Zn2+ shown as spheres and cysteines as sticks. The WT domain (top) with C446 (green) and C446G (red, bottom), demonstrating disrupted Zn2+ ligation of the C446G mutation also highlighted by a black arrowhead. (b) Solution structures of AIRE PHD1 (left, blue) and PHD2 (right, green) with the second Zn2+-binding region enlarged in each domain. Zn2+ shown as spheres and cysteines as yellow sticks; histidines in the first Zn2+-binding region are orange. The C311 and C446 residues (red), demonstrating their parallel position. (c) Family pedigree of individuals carrying an AIRE C446G mutation. Females are depicted as circles, males as squares. Diagonal lines indicate deceased individuals. (d) Chromatograms of DNA sequences around the C442G point mutation created. Nucleotide changes between the WT (top) and homozygous (bottom) sequence are highlighted by black triangles. (e) Representative flow cytometry plots of the TEC compartment of B6.Aire+/+, B6.Aire+/C442G, and B6.AireC442G/C442G mice. Numbers indicate the percentage of cells within each gate from the entire TEC compartment. (f) Frequencies of mTEChi (EpCAM+CD45−MHCIIhiLy51−/lo) and TEClo (EpCAM+CD45−MHCIIlo/midLy51−/lo) of B6.Aire+/+, B6.Aire+/C442G and B6.AireC442G/C442G mice. Frequencies are presented as the percentage from the average frequency of all WT animals within a given experiment. Data from two combined experiments with a total of five to eight mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. ***, P < 0.001 from the relevant WT littermate control. (g) Representative flow cytometry plots of T reg cells (CD4+CD8–CD25+Foxp3+) from thymi of B6.Aire+/+, B6.Aire+/C442G, and B6.AireC442G/C442G mice. Numbers indicate the percentage of cells within the gate from the CD4+ single-positive population. (h) Frequency and absolute counts of T reg cells (CD4+CD8–CD25+Foxp3+) in the thymi of B6.Aire+/+, B6.Aire+/C442G and B6.AireC442G/C442G mice. Both frequencies and absolute counts are presented as a percentage from the average frequency or count of all WT animals within a given experiment. Data from three combined experiments with a total of 6–12 mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. ***, P < 0.001 from the relevant WT littermate control. tTreg, thymic T reg. (i) Dendrogram and heatmap of Pearson correlation coefficient between the different mTEChi bulk RNAseq samples based on total gene expression values. (j) FC/FC plots of mTEChi gene expression comparing B6.Aire+/+/B6.Aire+/C442G (left, y axis) to B6.Aire+/+/B6.Aire−/− (x axis), or B6.Aire+/+/B6.AireC442G/C442G (right, y axis) to B6.Aire+/+/B6.Aire−/− (x axis). All genes are depicted by black dots along with a trendline for the best linear fit, and R2 is indicated in each plot. A signature of Aire-dependent TRA genes, which are down-regulated by at least 10-fold in B6.Aire−/− (based on a dataset from Sansom et al., 2014), is superimposed using red dots. (k) Frequency of AIRE+ mTEChi with significant colocalization (i.e., bright detail similarity >1.5) of AIRE and PML in B6.Aire+/+, B6.Aire+/C442G, and B6.AireC442G/C442G littermate mice. Data from four to six mice per group from two independent experiments, analyzed by one-way ANOVA, are represented as mean ± SEM. **, P = 0.0089 from the relevant WT littermate control. (l) Representative ImageStream snapshots of AIRE and PML colocalization in B6.Aire+/+, B6.Aire+/C442G, and B6.AireC442G/C442G littermate mice. BF, bright field. (m and n) Histogram for ChIPseq tag density for AIRE in B6.Aire+/+ and B6.Aire+/C442G mTEChi superenhancer regions defined in Bansal et al. (2017) (m) and AIRE-dependent TRA genes (n; as in j). TSS, transcription start site; TES, transcription end site.
To experimentally establish whether C446G is indeed a dominant-negative mutation, we generated another mouse model (AIRE C442G) using the CRISPR/Cas9 system. Although we initially tried to generate these mice directly on the NOD background, after multiple failed attempts to do so, we chose to use the C57BL/6 background (Fig. 5 d).
As expected, the overall thymic morphology in these mutants remained unaffected (Fig. S4 e). However, a typical increase in mTEChi frequency and absolute numbers (Fig. 5, e and f; and Fig. S4 f), as well as a decrease in the frequency and absolute counts of T reg cells in the thymus, were observed in the heterozygous Aire+/C442G mice (Fig. 5, g and h), similar to that found in Aire+/C313Y mice (Fig. 1, d and e; and Fig. S4 g). Other thymocyte populations remained unchanged in numbers, while some population frequencies were slightly altered (Fig. S4 g). Bulk transcriptomic analysis of the mTEChi compartment in B6.Aire+/C442G mice resulted in marked segregation between heterozygous mutants and their WT counterparts, and showed a large overlap in global gene expression between Aire+/C442G and the homozygous AireC442G/C442G mTEChi (Fig. 5 i). As in the case of the Aire+/C313Y mice, this differential clustering can be attributed to a specific reduction in AIRE-dependent TRA expression in mTEChi of Aire+/C442G mice as demonstrated in the FC/FC plot between Aire+/+ and Aire+/C442G in comparison to Aire+/+ and Aire−/− (Fig. 5 j), while AIRE-independent TRA genes remain unchanged (Fig. S4 h). The effect of Aire+/C442G on gene expression, however, is smaller compared with that of AireC442G/C442G, as seen by the reduced slope of the diagonal distribution and reduced correlation (R2) between gene expression profiles of Aire+/C442G and Aire−/− compared with that of AireC442G/C442G and Aire−/−. Therefore, the impact of the Aire+/C442G mutation on TRA expression is quantitatively smaller than that of Aire+/C313Y, suggesting that different mutations possess different dominant-negative capacities.
The relatively milder impact of C442G mutation on TRA expression was also mirrored by a relatively mild autoimmune phenotype. Specifically, while virtually all Aire−/− mice on C57Bl/6 background present with retinal degradation and autoimmune prostatitis (Jiang et al., 2005), retinal degeneration was observed only in 1 out of 21 aged Aire+/C442G mice examined; however, autoantibodies directed to the eye were seen in ∼50% of aged Aire+/C442G mice (Fig. S4 i). Furthermore, pronounced prostate infiltrates supporting a dominant-negative effect of the C442G PHD2 mutation were seen in the majority of aged Aire+/C442G male mice (Fig. S4 j).
Interestingly, unlike the AIREC313Y, the AIREC442G mutant was not sequestered to PML bodies (Fig. 5, k and l), suggesting that it must exert its dominant-negative effect by another mechanism. Therefore, to gain additional insights into its association with chromatin, we performed AIRE ChIPseq analysis on mTEChi isolated from either Aire+/+ or Aire+/C442G mice. A Spearman correlation coefficient between individual samples of 0.73–0.81, as well as 0.73 between the current findings and a previously published dataset using B6.Aire+/+ mTEChi (Bansal et al., 2017) was observed. Moreover, subsequent bioinformatic analysis revealed an average of 25,836 AIRE binding sites in the genome of Aire+/+ mTEChi compared with an average of 49,994 in Aire+/C442G mTEChi (Table S2; and Fig. S4, k and l). In contrast to the Aire+/C313Y ChIPseq described above, we observed higher read coverage of most AIREC442G peaks compared with those in Aire+/+ (Fig. S4 k). Moreover, differential binding analysis of all peaks revealed higher read concentration and higher enrichment of binding sites for Aire+/C442G compared with Aire+/+ (Fig. S4 l). Correspondingly, on superenhancer regions, Aire+/C442G showed increased (rather than decreased) binding capacity (Fig. 5 m and Fig. S4 m). However, AIREC442G showed a diminished capacity to bind its target TRA genes (Fig. 5 n), while an increased occupancy of AIREC442G was seen at AIRE-independent TRA loci (Fig. S4 n). These findings further support the divergent effects of various AIRE mutations, which may stem from their location in different domains. These results also suggest that while the PHD1 domain is critical for AIRE’s binding to the enhancers, the PHD2 domain may be critical for the subsequent looping of the enhancer regions with the corresponding target loci.
Dominant and recessive AIRE mutations differentially affect AIRE protein expression
To gain further insight into the differential effect of the described mutations on AIRE’s activity, we examined AIRE expression in the mice strains created. To this end, we performed SDS-PAGE on EpCAM-enriched stroma from all mutants including Aire−/− followed by Western blot (WB) using a polyclonal antibody against the SAND domain, enabling us to detect AIRE in all mutants studied (Fig. 6 a). As expected, AIRE protein was clearly detectable in EpCAM-enriched stroma of Aire+/+, while it was absent in Aire−/− mice. Interestingly, a striking difference was seen in AIRE expression between recessive and dominant-negative mutants. Specifically, in AireC313X/C313X mice, AIRE protein was completely absent, despite the assumption that mutations with such PTC (e.g., the Finnish mutation R257X) result in a nonfunctional truncated protein (Oftedal et al., 2015). The absence of a truncated protein could not be attributed to proteasomal degradation as incubation of EpCAM-enriched stroma of AireC313X/C313X mice with the proteasome inhibitor MG132 did not result in accumulation of such a truncated AIRE protein (Fig. 6 b). However, close examination of our bulk RNAseq data and subsequent validation by quantitative PCR (qPCR) revealed elevation of several genes (e.g., Dcp1a, Smg7, and Upf3a) related to the nonsense-mediated decay (NMD) pathway in AireC313X/C313X mice (Fig. 6 c). Furthermore, AIRE mRNA stability in both nuclear and cytosolic fractions was assessed by treating EpCAM-enriched stroma of AireC313X/C313X mice and WT littermates with the RNA Pol II poison actinomycin-D (ActD) for different time periods (Fig. S5 a). Interestingly, AIRE mRNA was more abundant in AireC313X/C313X compared with Aire+/+ mice at all time points and was more abundant in the nucleus compared with the cytosol (Fig. 6 d). While AIRE mRNA levels decreased at a similar rate in the nuclear fraction of both AireC313X/C313X and Aire+/+ mTECs, the mRNA was degraded more rapidly in the cytosol of AireC313X/C313X compared with Aire+/+ mTECs following ActD treatment. These data therefore suggest that while the AIRE C313X transcript is expressed at higher levels in the nucleus, it undergoes swift NMD in the cytosol (Fig. 6, e and f) without yielding any protein product (Fig. 6 a).
Figure 6.
Dominant and recessive AIRE mutations differentially affect AIRE protein expression. (a) WBs from EpCAM-enriched stroma of all homozygous mutants created in this study in addition to Aire−/−, compared with WT littermates and probed with an α-AIRE SAND antibody. (b) WBs from EpCAM-enriched stroma of B6.Aire+/+ and B6.AireC313X/C313X mice treated with the proteasomal inhibitor MG132 or DMSO and probed with α-AIRE SAND antibody. (c) Relative mRNA expression of NMD-related genes from sorted mTEChi from B6.Aire+/+ and B6.AireC313X/C313X littermates. Data from three mice per group, analyzed by Student’s t test, are represented as mean ± SEM. *, P < 0.05; **, P < 0.01 from WT littermates. (d) Histograms of relative AIRE mRNA abundance in the nucleus (Nuc) versus cytosol (Cyto) from EpCAM-enriched stroma of B6.Aire+/+ and B6.AireC313X/C313X littermates treated for 0 h, 0.5 h, 1 h, or 2 h with ActD and assessed by qPCR. Representative figure from one of two independent experiments. (e and f) Relative abundance of nuclear (e) or cytosolic (f) AIRE mRNA in B6.Aire+/+ and B6.AireC313X/C313X EpCAM-enriched stroma following incubation with ActD for the time points indicated and assessed by qPCR using the ΔΔCt method, normalized to Hprt. Data are representative of two independent experiments. (g–i) WBs from EpCAM-enriched stroma of B6.Aire+/+ and B6.AireY86C/Y86C mice treated with protease inhibitor (PI) or DMSO and probed with α-AIRE SAND antibody (g). AIRE is indicated by a black arrow. WB representative of two independent experiments. Followed by quantification of AIRE protein FC following treatment with protease inhibitors in B6.Aire+/+ and B6.AireY86C/Y86C (h). AIRE protein levels per sample were first normalized to GAPDH, and then FC between PI treatment and DMSO was calculated. Lines between samples indicate samples from the same independent experiment. The relative increase in AIRE protein levels compared with the relevant WT control is shown in panel i. Data from two independent experiments, analyzed by Student’s t test, are represented as mean ± SEM. **, P < 0.01 from WT. (j) Representative ImageStream snapshots of AIRE+ mTECs from both AireC313Y and AireC442G heterozygous and homozygous mice. BF, bright field. (k–m) Frequencies of AIRE+ mTEChi (EpCAM+CD45–MHCIIhiLy51–/loAire+; k), AIRE mean fluorescence intensity (MFI; k), and AIRE mRNA levels (based on normalized UMI count from bulk RNAseq data; m) in all mice created in this study in addition to Aire−/−. Frequencies, MFI, and AIRE mRNA levels are calculated as a percentage from the average frequency, MFI, or normalized UMI count of all WT animals within a given experiment. Each mouse strain was examined separately. Data from two to six mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from the relevant WT littermate controls. NOD.AireC313Y/C313Y mice were used for assessment of frequency and MFI, while NOD.AireC313Y/– mice were used in RNAseq, from which normalized UMI counts were extracted. Data for AIRE+ mTEChi frequencies and AIRE MFI are representative of two independent experiments.
Figure S5.
Dominant and recessive AIRE mutations differentially affect AIRE protein expression, and AIRE directly regulates its own expression. (a) WBs from nuclear and cytosolic fractions of EpCAM-enriched stroma from B6.Aire+/+ and B6.AireC313X/C313X mutants created in this study, probed with an α-H2B and α-GAPDH antibodies to verify proper cell fractionation. (b) Heatmap of protease genes expression of >150 UMIs in mTEChi from B6.Aire+/+ mice, which are also up-regulated in B6.AireY86C/Y86C mice. (c) WBs from EpCAM-enriched stroma from dominant-negative NOD.Aire+/C313Y (left) and B6.Aire+/C442G (right) created in this study probed with an α-AIRE SAND and α-GAPDH antibodies. (d) Representative flow cytometry plots of Aire+ mTEChi (EpCAM+CD45–MHCIIhiLy51–/loAire+) from Aire Y86C, C313X, V303M, C313Y, and C442G mouse strains created, in addition to Aire−/− on both B6 and NOD backgrounds. Numbers indicate the percentage of cells within each gate from the mTEChi population. (e) BDI of AIRE intracellular stain from ImageStream analysis of NOD.Aire+/C313Y (left) and B6.Aire+/C442G (right) mice. BDI is calculated as the intensity of localized bright spots that are 3 pixels in radius or less within the masked area in the image, with the local background around the spots removed. Data from four to seven mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. **, P < 0.01, from the relevant WT littermate controls. Data are representative of two independent experiments for each strain. (f) MFI of AIRE intracellular stain from flow cytometric analysis of NOD.Aire+/C313Y (left) and B6.Aire+/C442G (right) mice. Data from three to six mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. ***, P < 0.001, from the relevant WT littermate controls. Data are representative of two independent experiments for each strain. (g) Representative flow cytometry plots of Aire.GFP+ mTEChi (EpCAM+CD45–MHCIIhiLy51–/loAire.GFP+) from Adig.Aire+/+, Adig.Aire+/−, and Adig.Aire−/− mice, as well as Adig.Aire+/+, Adig.Aire+/Y86C, and Adig.AireY86C/Y86C mice. Numbers indicate the percentage of cells within each gate from the mTEChi population. (h) Pairwise correlation (Spearman’s) showing overall similarity between read coverage within the same genomic regions in the ChIP or ATAC samples. Each dot is a genomic region, axes show the number of fragments in natural log scale, and Spearman coefficient is denoted on the graph.
A reduction in AIRE protein expression was also apparent in AireY86C/Y86C mice (Fig. 6 a). Several previous publications showed that overexpression of Y85C or other CARD mutations in transfected cells impairs its speckled nuclear localization and results in diffuse nuclear staining (Halonen et al., 2004; Huoh et al., 2020; Oftedal et al., 2015; Ramsey et al., 2002b). Y85C was also reported to have more rapid decay than other AIRE mutants in vitro (Ramsey et al., 2002a). It was also previously suggested that the recessive nature of Y85C is due to its inability to form homo-oligomers, which are necessary for AIRE’s function (Oftedal et al., 2015). Interestingly, treating EpCAM-enriched stroma of AireY86C/Y86C mice with protease inhibitors revealed that the Y86C mutant is more prone to proteolytic degradation than its WT counterpart, as evident by the presence of a higher proportion of AIRE protein following short treatment with protease inhibitors in AireY86C/Y86C mice compared with their Aire+/+ littermates (Fig. 6, g–i). Increased gene expression of various proteases, proteasomal subunits, and their associated genes in mTEChi, specifically in AireY86C/Y86C mice, further supports increased proteolytic degradation of AIRE in these mice (Fig. S5 b). Interestingly, Ctss and Ctsh, encoding for cathepsin S and H, respectively, stand out in this list as both are cysteine proteases, which would likely be active against the elevated levels of exposed cysteine residues due to the tyrosine to cysteine substitution in Y86C. Thus, our data suggest that the observed instability and increased proteolytic degradation of the Y86C mutant may stem from its inability to form functional homo-oligomers. Importantly, contrary to the recessive mutants, AIRE protein was readily detected at the expected size in AireV303M/V303M, AireC313Y/– and AireC442G/C442G mice (Fig. 6 a), as well as in both dominant-negative mutants (Fig. S5 c) and with the characteristic speckled form in both dominant-negative heterozygotes as well as in AireC313Y/C313Y (Fig. 6 j). The apparent increase in AIRE intensity for the AireC313Y/– and AireC442G/C442G mutants in the WB likely reflected the relative increase of mTEChi within the EpCAM-enriched fraction. However, clear defects in AIRE subnuclear localization were apparent in AireC442G/C442G, as only very few large speckles could be detected in AIRE-expressing cells.
To better quantify the differences in AIRE protein expression, we performed flow-cytometric analysis of the mTEChi compartment of the individual mutant strains, using the monoclonal 5H12 antibody directed at AIRE’s C terminus (20 final amino acids; Hubert et al., 2009). These analyses showed that mice harboring the heterozygous recessive mutations Aire+/Y86C and Aire+/C313X or the Aire+/V303M mutation have either no change or a slight decrease in the frequency of AIRE+ cells and/or in the intensity of AIRE intracellular staining compared with Aire+/+ littermates (Fig. 6, k and l; and Fig. S5 d). As expected, no AIRE protein was detected in AireC313X/C313X mice as the PTC in these mice is located in exon 8 (out of 14). Surprisingly, no AIRE+ mTEChi cells were also detected in AireY86C/Y86C mice, suggesting that the residual traces of AIRE Y86C seen in the WB (Fig. 5 a) may have a different conformation (e.g., nonfunctional monomers) that cannot be detected by this specific antibody clone. Interestingly, a significant increase in the frequency of AIRE+ mTEChi was found in both Aire+/C313Y and Aire+/C442G heterozygous mice (Fig. 6 k and Fig. S5 d). This elevated frequency is coupled with increased intensity of AIRE intracellular staining in both strains of heterozygous dominant mutants as assessed by both flow cytometry and ImageStream (Fig. 6 l; and Fig. S5, e and f), suggesting that AIRE is present with augmented stability or is more readily transcribed/translated in Aire+/C313Y and Aire+/C442G mice. Indeed, an increase in AIRE mRNA transcripts is evident in all homozygous mutants described above, as well as in both heterozygous dominant mutants (Fig. 6 m), suggesting that functional AIRE is necessary to auto-regulate its own transcription.
Collectively, these data demonstrate that while recessive mutations result in a lack of AIRE protein expression, the dominant mutations surprisingly augment the expression of the AIRE protein, suggesting that the expression or absence of expression of the given AIRE mutant is a key determinant of whether the mutation will be dominant or recessive.
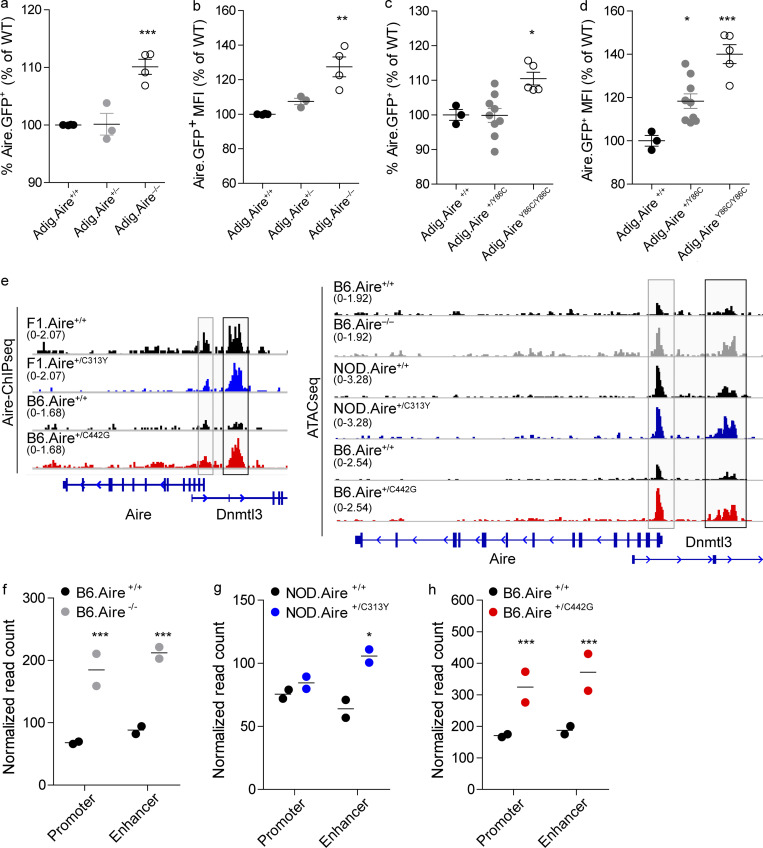
AIRE directly regulates its own expression
Further validation of our hypothesis that AIRE may regulate its own expression was provided by analysis of Aire-IGRP-GFP (Adig) reporter mice (Gardner et al., 2008), in which a GFP reporter is fused to the pancreatic self-antigen Igrp and expressed under the Aire promoter. Specifically, B6.Adig mice were bred with B6.Aire+/− mice to produce Adig.Aire+/+, Adig.Aire+/−, and Adig.Aire−/− mice, which harbor a sole copy of the Adig reporter. Analysis of Adig.Aire−/− mice showed a significantly higher frequency of GFP+ mTEChi cells (Fig. 7 a and Fig. S5 g) with higher GFP fluorescence intensity (Fig. 7 b) compared with Adig.Aire+/+ and Adig.Aire+/− mTEChi, supporting over-activation of the proximal Aire regulatory locus. Comparable results were obtained when Adig.AireY86C/Y86C mice were examined in a similar fashion (Fig. 7, c and d; and Fig. S5 g). Based on these data, we hypothesized that AIRE may modulate its own expression through direct binding to its own cis-regulatory elements. To experimentally test this, we employed our AIRE ChIPseq datasets in mTEChi and examined AIRE protein occupancy at its own locus. Indeed, we observed significant enrichment of AIRE at its own promoter and proximal enhancer (i.e., CNS1; Haljasorg et al., 2015; LaFlam et al., 2015; Fig. 7 e), further supporting an autoregulatory role for AIRE. Interestingly, both C313Y and C442G dominant mutations bound these regulatory regions (Fig. 7 e), indicating that altered fold of either PHD domain does not impinge on AIRE binding to its own locus. To further validate whether AIRE is indeed able to limit the accessibility of its own regulatory elements, we performed assay of transposase-accessible chromatin following sequencing (ATACseq) analysis of mTEChi isolated from WT, Aire−/−, Aire+/C313Y, or Aire+/C442G heterozygous mice and found high correlations between our ChIPseq and ATACseq data (Fig. S5 h). Indeed, assessment of chromatin accessibility of these loci revealed increased accessibility of both the Aire proximal promoter and enhancer in B6.Aire−/− mTEChi (Fig. 7, e and f) and similarly increased accessibility of the Aire proximal enhancer in Aire+/C313Y (Fig. 7, e and g) and both the Aire promoter and proximal enhancer in Aire+/C442G mTEChi (Fig. 7, e and h). Taken together, these findings strongly support the existence of a negative feedback loop in which AIRE aids other previously identified transcription factors (Herzig et al., 2017) in limiting accessibility to these loci to reduce its own transcription. However, while both dominant-negative mutants were able to bind these regulatory loci, their ability to induce changes in the accessibility of these loci is limited.
Figure 7.
AIRE directly regulates its own expression. (a–d) Frequencies (a and c) and MFI (b and d) of Aire.GFP+ mTEChi (EpCAM+CD45−MHCIIhiLy51−/loAire.GFP+) in Adig.Aire+/+, Adig.Aire+/−, and Adig.Aire−/− (a and b) or Adig.Aire+/+, Adig.Aire+/Y86C, and Adig.AireY86C/Y86C (c and d) mice. Mice were bred such that only one of the parents had a single Adig allele. Each dot represents an individual mouse. Frequencies and MFI are calculated as a percentage from the average frequency or MFI of all WT animals within an experiment. Data from three to nine mice per group are analyzed by one-way ANOVA and are represented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from the relevant WT littermate control. Each mouse strain was evaluated separately. At least two independent experiments were conducted on each strain. See also Fig. S5 g. (e) Normalized AIRE ChIPseq profiles (left) at the Aire locus of F1.Aire+/+ (black), F1.Aire+/C313Y (blue), B6.Aire+/+ (black), and B6.Aire+/C442G (red) mTEChi, and normalized ATACseq profiles (right) at the Aire locus of Aire+/+ (black) and B6.Aire−/− (gray), Aire+/+ (black) and Aire+/C313Y (blue), and Aire+/+ (black) and Aire+/C442G (red) mTEChi. The Aire promoter is highlighted by a gray box, while the Aire proximal enhancer is highlighted by a black box. The range of normalized tag densities is indicated by the numbers in parentheses at the left of each track. (f–h) Normalized read count of individual ATACseq samples (n = 2 per group) at the Aire promoter and Aire enhancer for each of the mouse strains examined. Statistical significance (*, adjusted P < 0.05; **, adjusted P < 0.01; ***, adjusted P < 0.001) from the relevant WT control was determined by DiffBind.
In summary, these findings reveal another layer of complexity in the tight regulation of the Regulator and identify a novel mechanism by which AIRE modulates its own transcription by repressing chromatin accessibility of its proximal regulatory elements.
Discussion
While the main manifestations of APS-1 (i.e., hypoparathyroidism, adrenocortical insufficiency, and chronic mucocutaneous candidiasis) are well-defined and serve as hallmarks for its diagnosis, APS-1 patients typically suffer from a heterogeneous array of additional autoimmune components including enamel hypoplasia, enteropathy, premature ovarian insufficiency, type 1 diabetes, autoimmune hepatitis, and more (Husebye et al., 2018). In recent years, the heterogeneity involved in AIRE-induced autoimmunity has grown in complexity as several dominant-negative mutations in AIRE were reported (Abbott et al., 2018; Cetani et al., 2001; Oftedal et al., 2015). Patients harboring these monoallelic mutations often do not qualify for the clinical diagnosis of APS-1, as they present with more common organ-specific autoimmunity such as autoimmune thyroid disease, vitiligo, pernicious anemia, and type 1 diabetes. IFN-ω and -α autoantibodies, present in almost all APS-1 patients with biallelic mutations, are much less prevalent in those with monoallelic dominant mutations. These patients thus challenge the notion that AIRE-induced autoimmunity is only a result of recessive modes of inheritance and suggest that autoimmunity due to AIRE mutations is much more common than originally believed (Eriksson et al., 2021).
In general, dominant-negative mutations give rise to a dysfunctional protein, which in turn acts as a competitive inhibitor of the WT protein. Dominant-negative mutations are typically found in homo-oligomeric proteins, form abnormal oligomers (Barndt et al., 2000; Jacenko et al., 1993), and thereby inhibit protein–protein interactions required for normal protein function (Grasberger et al., 2005), including DNA binding of transcription factors (e.g., STAT3 and MEF2; Asou et al., 2003; Caldenhoven et al., 1996; Ornatsky et al., 1997) and receptor binding to natural ligands (e.g., FAS and IFNGR1; Jouanguy et al., 1999; Siegel et al., 2000). Given that AIRE was shown to exist in homo-dimeric or homo-tetrameric forms (Kumar et al., 2001), the existence of various dominant-negative AIRE mutants is not surprising.
Here we created several mouse models that bear either various known recessive or different putative dominant-negative AIRE patient mutations and have validated that the Aire+/C313Y monoallelic mutation operates in a dominant-negative manner, as it almost completely abrogates the expression of AIRE-dependent genes in mTECs and gives rise to a fatal autoimmune phenotype, similar to that in Aire−/− mice. However, the extent of autoimmunity was always milder than that observed in Aire−/− mice. This is well in line with data from human patients, showing that dominant monoallelic AIRE mutations typically present with milder autoimmune phenotypes that in many instances would not be classified as APS-1.
An open question that arose from studying autoimmune patients with dominant-negative mutations was whether these mutations are restricted only to the PHD1 and SAND domains or whether they could also extend to other AIRE domains. Based on spatial similarities with C311Y and previous in vitro data (Oftedal et al., 2015), we predicted that the C446G patient mutation in AIRE’s PHD2 domain shall exert an analogous dominant-negative effect. Indeed, our analysis of a newly established Aire+/C442G mouse model supported this assumption, as it resulted in impaired expression of AIRE-dependent TRA genes in mTECs, reduced frequency of Foxp3+ T reg cells in the thymus, and mild autoimmunity on the B6 background. Interestingly, the impact of the Aire+/C442G mutation on the expression of AIRE-dependent TRA genes in mTECs was reduced compared with that of Aire+/C313Y, suggesting that different monoallelic mutations possess different dominant-negative capacities and correspond to the respective phenotypes in humans. This has important clinical implications, as the scope of autoimmune cases due to dominant-negative mutations in AIRE may be broader and more heterogeneous than previously thought and warrants further investigation of relevant family members of APS-1 patients carrying this mutation.
Interestingly, AIREC313Y and the AIREC442G mutations seem to differ in their modus operandi. First, the AIREC313Y as well as the AIREC442G homozygous mutants (but not the AIREC442G heterozygous mutant) show aberrant subnuclear localization, as AIREC313Y mutants are sequestered into PML bodies, while the homozygous AIREC442G mutants appear in very few enlarged nuclear speckles. These differences in subnuclear localization likely impact their stability and/or turnover. Historically, PML bodies were considered to be devoid of DNA; however, recent studies have found that PML bodies can physically interact with chromatin, and in untreated cells, PML bodies are surrounded by chromatin, which is important for the positional stability of these membrane-less domains (Eskiw et al., 2004). Furthermore, there is evidence that PML bodies can contain chromatin within them (Lang et al., 2010; Luciani et al., 2006), and ChIPseq of PML has been published (Delbarre et al., 2017). Therefore, the association of Aire+/C313Y with PML seen in our imaging flow cytometry experiments (Fig. 4, a and b) does not obviate its ability to bind DNA, as seen in our ChIPseq findings (Fig. 4, c–e), though it might change the chromatin landscape to which a proportion of Aire+/C313Y cells have accessibility. In support of the role of the AIRE PHD1 domain as a histone reader (Koh et al., 2008; Org et al., 2008), the AIREC313Y is not as efficient in binding both AIRE-dependent TRA loci and superenhancer regions compared with Aire+/+. In contrast, the AIREC442G mutant showed increased (rather than decreased) capacity to bind superenhancer regions, while it had diminished capacity to bind its target TRA genes (Fig. 8). These results argue for different roles of both of AIRE’s PHD domains in transcriptional regulation and suggest that while the PHD1 domain is critical for AIRE’s binding to the enhancers, the PHD2 domain may facilitate the subsequent looping of the enhancer regions with the corresponding target loci.
Figure 8.
The impact of different AIRE mutations on AIRE expression and activity. In Aire+/+, Aire+/−, or Aire+/recessive mTEChi, AIRE takes part in maintaining an equilibrium of its own expression. Specifically, AIRE tetramers bind the accessible Aire proximal enhancer and aid other factors at the locus to limit Aire enhancer accessibility, thereby limiting AIRE transcription. When AIRE levels are low or absent, as in Airerecessive/recessive or Aire−/− mTEChi, AIRE cannot affect the accessibility of the Aire proximal enhancer, which remains open and active, leading to the production of excessive amounts of AIRE mRNA. The nature of the recessive mutation will determine whether the mRNA will be degraded directly (e.g., mutations with PTCs such as C313X, frameshift), or whether it will be translated, but then degradation will occur at the protein level (e.g., mutations that render AIRE unable to oligomerize such as Y86C). Similarly, in Aire+/dominant mTEChi, the Aire proximal enhancer is maintained open and active as dysfunctional AIRE complexes binding the locus cannot partake in limiting the enhancer accessibility, leading to accumulation of increased amounts of mutated AIRE mRNA. This is also mirrored by increased levels of AIRE protein complexes, which are, however, dysfunctional and incapable of binding AIRE target genes (e.g., C442G) and/or superenhancers (C313Y).
Contrary to the robust phenotype observed in mice carrying a single Aire+/C313Y or Aire+/C442G mutation, we were surprised to find that mice carrying a Aire+/V303M mutation exhibited no phenotype. Although the V301M mutation was previously suggested to have an incomplete dominant-negative capacity in transfected cell lines (Oftedal et al., 2015), our findings argue that it represents a rather benign variant with a relatively high prevalence among the general population (0.00089). Although this prediction is in line with studies demonstrating that substitution of valine for methionine results in loss of function in various genes such as the androgen receptor (Kazemi-Esfarjani et al., 1993), other studies have demonstrated that the V301M substitution in AIRE does not result in any change to the structure or to the stability of AIRE’s PHD1 domain (Gaetani et al., 2012; Koh et al., 2008), and thus would not be expected to impact the domain or AIRE function. Furthermore, these findings explain the lack of reported APS-1 cases with V301M homozygosity or even V301M in combination with other mutations in APS-1 patients.
The creation and comparison of the phenotypes of several dominant-negative and recessive AIRE point mutants allowed us to understand why certain mutations are recessive while others are dominant-negative, in addition to gaining critical insight into the regulation of AIRE itself. We found that the main factor differentiating between the dominant-negative and recessive mutations was AIRE expression or lack thereof (Fig. 8). While both C313Y and C442G heterozygous mutations resulted in a stable (but functionally inactive) protein, which could readily be detected by WB or by flow cytometry analyses, homozygous recessive mutations resulted in very little (Y86C) or no AIRE protein being expressed (C313X). Therefore, these results challenge conclusions of several previous publications (based on transfected cell lines), which reported that recessive AIRE mutations give rise to either truncated or mislocalized AIRE proteins (Halonen et al., 2004; Huoh et al., 2020; Oftedal et al., 2015; Ramsey et al., 2002b). Therefore, our results suggest that the built-in physiological regulatory mechanisms that are employed in vivo (e.g., NMD, proteolytic degradation) might not occur under artificial in vitro conditions, when cell lines are transfected with plasmids lacking introns. These important differences underscore the caution that should be taken when generalizing findings made in vitro. Furthermore, our findings also highlight differences that can occur between a heterozygous dominant-negative mutation and its respective homozygous counterpart, as seen in the case of AIREC442G, where differences in AIRE’s nuclear localization, appearance, and intensity per cell are apparent. Our findings further explain why different mutations on the same amino acid (e.g., C313) can give rise to either a dominant-negative or a recessive mutation. Specifically, while the C313Y mutation resulted in a stable though nonfunctional protein, the C313X mutation gave rise to a PTC, which triggered NMD mechanisms of AIRE mRNA, ultimately leading to the lack of AIRE protein in these mice.
Not only was AIRE expressed in the dominant-negative mutants, it was also expressed in a higher percentage of mTEChi, and with higher expression per cell, suggesting that the mutations either stabilize the protein or induce increased transcription of AIRE. Indeed, we found that in all mutants with nonfunctional AIRE, AIRE mRNA was significantly increased, implicating AIRE in autoregulation (Fig. 8). This notion is further supported by additional data demonstrating that AIRE physically binds its own proximal promoter and enhancer, even when the histone-reading PHD1 domain is mutated in Aire+/C313Y. Moreover, ATACseq analysis on mTEChi from both C313Y and C442G heterozygous mutants shows increased accessibility of this enhancer element compared with WT mTEChi, indicating that while AIRE from both dominant-negative mutants is capable of binding this locus, its altered folding does not allow proper chromatin remodeling of the locus, further supporting an autoregulatory role for AIRE in limiting accessibility to its own enhancer elements, which would ultimately limit its own transcription. This is in accordance with a previous report of AIRE’s intrinsic repressive effect on chromatin accessibility (Koh et al., 2018). Use of such negative autoregulatory loops was shown to be crucial for maintaining an optimal concentration of transcription factors as seen in the case of OCT4 (Pan et al., 2006) and BRCA1 (De Siervi et al., 2010). Such autoregulatory feedback loops are highly conserved in vertebrates (Kiełbasa and Vingron, 2008) and have previously been shown to significantly reduce the response time in transcription networks (Rosenfeld et al., 2002), which in the case of AIRE would allow better and tighter control of promiscuous gene expression.
In conclusion, our data elucidate why some AIRE mutations are recessive while others are dominant and bring insight into AIRE’s modus operandi by identifying a novel autoregulatory mechanism by which AIRE negatively modulates its own expression, through direct binding to its own cis-regulatory elements.
Materials and methods
Mice
NOD/ShiLtJ (#001976) and NOD Scid Gamma (NSG; #005557) mice were purchased from Jackson Laboratories. B6.Aire−/− and NOD.Aire−/− were kindly provided by Diane Mathis and Christophe Benoist (Harvard University, Boston, MA). Adig (Gardner et al., 2008) mice were kindly provided by Mark S. Anderson (University of California, San Francisco, CA). B6.AireY86C, B6.AireC313X, B6.AireC442G, NOD.AireV303M, and NOD.AireC313Y mice were generated at the transgenic facility at the Weizmann Institute of Science, using CRISPR/Cas9 genome editing in isolated one-cell embryos from C57Bl/6 or NOD mice, respectively. Generating these mice on a NOD background was attempted for most strains, but was not always successful even after several attempts as the number of oocytes successfully harvested and fertilized following in vitro fertilization was much lower compared with C57Bl/6. Therefore, the C57Bl/6 background was used to generate some of the mutations. Genotyping of mice with point mutations was conducted using custom TaqMan SNP Genotyping assays (Table S4). The B6.AireC442G mice were genotyped by Sanger sequencing of the relevant locus as an adequate TaqMan SNP Genotyping assay could not be generated.
As NOD mice have smaller thymi yielding fewer TECs, NOD.Aire+/C313Y males were bred with WT C57Bl/6 females (Envigo) for ChIPseq experiments. F1 progeny were used for sorting mTEChi to obtain more cells per mouse.
The early onset of autoimmunity in NOD.Aire+/C313Y mice hampered their ability to breed with other NOD.Aire+/C313Y mice. Therefore, WT NOD/ShiLtJ mice or NOD.Aire+/− mice were used to produce both NOD.Aire+/C313Y and NOD.AireC313Y/– mice. All mice were maintained and bred under specific pathogen–free conditions at the Weizmann Institute of Science’s animal facility and were provided with standard rodent chow and autoclaved water ad libitum. 4–7-wk-old mice were used in all experiments unless stated otherwise, and all handling and experimentation were conducted in accordance to the guidelines of the Weizmann Institute of Science Institutional Animal Care and Use Committee (#10060119-2). All control mice used were littermates of the relevant mutant strain.
Human subjects
Patients were recruited from the Norwegian National Registry of Autoimmune Diseases. The study was approved by the Regional Committee for Medical and Health Research Ethics (2009/2555), and informed consent was provided by all subjects.
Modeling AIRE domain structure
Protein structures of AIRE mutations in the CARD, PHD1, and PHD2 domains were generated using PyMOL (http://www.pymol.org). For the PHD1 and PHD2 domains, the previously published nuclear magnetic resonance structures 1XWH (Bottomley et al., 2005) and 2LRI (Gaetani et al., 2012) were used as templates for modeling. For the CARD domain mutations, homology modeling was performed using the first 104 residues of the AIRE protein sequence with the Phyre2 homology modeler using the intensive mode (Kelley et al., 2015). The ensuing structure was modeled at >90% confidence for 93% of residues, using template structures from CARD9 and NOD1. These AIRE domain structures were subsequently modified using the mutagenesis feature within PyMOL, and then processed using the clean command on residues in close proximity to the modification. For the purpose of comparing the location and orientation of the cysteines C311 in PHD1 and C446 in PHD2, pair fitting was performed using the four cysteines within the second Zn+ binding region of both domains.
Isolation of TECs and thymocytes, flow cytometry and ImageStream analysis, sorting, and data processing
TECs
Thymi were dissociated by enzymatic digestion using 16.6 µg/ml Liberase TH (LIBTH-RO; #540113; Roche) and 10 µg/ml DNase in RPMI at 37°C until complete digestion. The single-cell suspension was then filtered through a 52-µm mesh filter and resolved on a Percoll gradient. To this end, the single-cell suspension was washed and resuspended in 1.115 g/ml isotonic Percoll (P1644; Sigma-Aldrich), topped by one layer of isotonic 1.065 g/ml Percoll and one layer of 1× PBS. The Percoll gradient was centrifuged at 2,700 rpm at 4°C with no break for 30 min. Stromal cells, found between the 1× PBS layer and the 1.065 g/ml Percoll layer, were collected and washed with MACS buffer (2% FBS with 5 mM EDTA, pH 8.0, in 1× PBS) followed by centrifugation at 340 g for 5 min at 4°C. Cells were then stained with specific antibodies.
Thymocytes and T reg cells
Thymi were collected in 1× PBS and kept on ice. Single-cell suspensions were prepared by mechanical dissociation of the thymi through a 40-µm strainer using a syringe plunger.
The following antibodies were used for surface immunostaining of thymic stromal cell suspensions: EpCAM APC (118214; Biolegend), EpCAM APC-Cy7 (118218; Biolegend), CD45 FITC (103108; Biolegend), CD45 PE-Cy7 (103114; Biolegend), CD45 PerCP-Cy5.5 (103132; Biolegend), Ly51 PE (108308; Biolegend), Ly51 PE-Cy7 (108314; Biolegend), CD80 Pacific Blue (104724; Biolegend), IA-IE Pacific Blue (107620; Biolegend), and IA-IE APC (107614; Biolegend). IAg7 was a kind gift from Diane Mathis and Christophe Benoist and was conjugated to Pacific Blue or APC. The following antibodies were used for membranal immunostaining of thymocytes and T reg cells: CD4 PE-Cy7 (100422; Biolegend), CD8a APC (100712; Biolegend), and CD25 PE (101904; Biolegend). DAPI (D9542; Sigma-Aldrich) or viability dye eF506 (65–0866-14; eBioscience) was used for live/dead cell discrimination. For intracellular staining of AIRE, Foxp3, or PML, cells labeled for membrane antigens were washed and then fixed with the True-Nuclear Transcription Factor Buffer set (424401; Biolegend) as per the manufacturer’s instructions, followed by AIRE AF488 (53–5934-82; eBioscience) or Foxp3 AF488 (126406; Biolegend) staining. For PML staining, the ab190899 antibody (Abcam) was used, followed by secondary staining with goat anti-rabbit AF647 (111–605-144; Jackson ImmunoResearch). Following staining, cells were washed, resuspended in MACS buffer, filtered, and either analyzed on a BD FACS Canto II analyzer or sorted using a BD FACS Aria III cell sorter. Cell counts were determined using cell counting beads (ACBP-100-10; Spherotech). Data analysis was conducted using FlowJo software (Tree Star). For ImageStream experiments, cells were resuspended in 20–30 µl MACS buffer, and acquired and analyzed using IDEAS 6.2 software (Amnis Corp., part of Luminex). The ImageStreamX Mark II flow cytometer (Amnis Corp.) is equipped with four lasers: 405 nm (120 mW), 488 nm (100 mW), 561 nm (200 mW), and 642 nm (150 mW). Laser intensities were set to maximal values that do not saturate the camera. Cells were gated for single cells using the area (sum of pixels within a mask, in square microns) and aspect ratio (the minor axis divided by the major axis), and for focused cells using the gradient root mean square feature (measures the sharpness quality of an image by detecting large changes of pixel values in the image, computed using the average gradient of a pixel normalized for variations in intensity levels). Bright detail intensity (BDI) is calculated as the intensity of localized bright spots that are 3 pixels in radius or less within the masked area in the image, with the local background around the spots removed. Co-localization was quantified using the bright detail similarity feature (compares the small bright image detail of two images, calculated as the log-transformed Pearson’s correlation coefficient of the localized bright spots with a radius of 3 pixels or less within the masked area in the two input images).
RNA isolation, sequencing, and analysis
For the purpose of bulk RNAseq, cells were sorted into 50 µl of Lysis/Binding Buffer (A33562; Invitrogen) followed by RNA extraction using Dynabeads (61012; Invitrogen) according to the manufacturer’s instructions. Sequencing libraries were prepared according to the massively parallel single-cell RNAseq method (Jaitin et al., 2014) and sequenced on an Illumina NextSeq500 platform. Massively parallel single-cell RNAseq was analyzed using the user-friendly transcriptome analysis pipeline (Kohen et al., 2019). Briefly, raw reads were trimmed using cutadapt with the following parameters: -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -a “A{10}” -times 2 -u 3 -u -3 -q 20 -m 25. Reads were mapped to the genome (mm10, Refseq annotation) using STAR (v2.4.2a) with the parameters -alignEndsType EndToEnd, -outFilterMismatchNoverLmax 0.05, -twopassMode Basic, -alignSoftClipAtReferenceEnds No. The pipeline quantifies the 3′ of Refseq annotated genes (The 3′ region contains 1,000 bases upstream of the 3′ end and 100 bases downstream). Unique molecular identifier (UMI) counting was done after marking duplicates (in-house script) using HTSeq-count in union mode. Only reads with unique mapping were considered for further analysis, and genes having a minimum of five reads in at least one sample were considered. Gene expression levels were calculated and normalized using DESeq2 with the following parameters: betaPrior = True, cooksCutoff = FALSE, and independentFiltering = FALSE. Raw P values were adjusted for multiple testing, using the Benjamini–Hochberg procedure. Differentially expressed genes were selected with absolute FC (log2) ≥ 2 and adjusted P value ≤0.05.
For the purpose of gene expression by qPCR, cells were sorted into 50 μl of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, pH 8, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, and 0.1% sodium deoxycholate). 750 μl TRIreagent (T9424; Sigma-Aldrich) was added to lyse the cells, and RNA was extracted according to the manufacturer’s protocol. RNA was then reverse-transcribed using the High-Capacity cDNA Reverse-Transcription kit (4368814; Thermo Fisher Scientific) using random primers. The subsequent PCR analysis was performed with Fast SYBR Green Master Mix (4385614; Thermo Fisher Scientific) and specific primers (Dcp1a Fwd, 5′-TGTACACTTTCTGCCCCAAA-3′, Rev, 5′-CGGTATACAAATAACGTCCCTTCT-3′; Smg7 Fwd, 5′-CCAGGATGCCATTTGAGG-3′, Rev, 5′-GCAGCCAGACTCTTTAACAGG-3′; Upf3a Fwd, 5′-CAGGAAGACCGGCTGGTAG-3′, and Rev, 5′-TTGGCTCTTTATCACTGGTATCC-3′). All samples were analyzed on the Viia7 real-time PCR instrument (Life Technologies), and differential expression was calculated according to the change-in-cycling threshold (ΔΔCT) method and normalized to the housekeeping gene Hprt (Fwd, 5′-TGAAGAGCTACTGTAATGATCAGTCAA-3′, and Rev, 5′-AGCAAGCTTGCAACCTTAACCA-3′).
AIRE mRNA stability assay
For the assessment of AIRE mRNA stability, following Percoll enrichment of thymic stromal cells (see above), cells were further enriched for TECs by staining for EpCAM-APC (118214; Biolegend), followed by positive selection using anti-APC microbeads (130–090-855; Miltenyi Biotec). The enriched cells of each genotype were pooled, counted, and then washed in MACS buffer (340 g, 5 min, 4°C). EpCAM-enriched cells were treated with RPMI medium (1% L-glutamine, 1% pen-strep, and 10% FBS) supplemented with 10 µg/ml ActD (A9415; Sigma-Aldrich; diluted in DMSO, D2438; Sigma-Aldrich). Cells were then incubated at 37°C with 5% CO2 for 0 h, 1/2 h, 1 h, and 2 h, at which time they were harvested by gently scraping the dish and collecting the cell suspension. Cells were washed in cold 1× PBS and then transferred to a chilled microtube and washed again in 1 ml 1× PBS. To separate the cytoplasmic fraction from the nuclear fraction, the cell pellet was then resuspended in 100 μl hypotonic buffer (20 mM Tris-HCl, pH 7.4, 10 mM NaCl, and 3 mM MgCl2) supplemented with RNase inhibitor (N8080119; Thermo Fisher Scientific; 1:50). Cells were incubated for 15 min on ice. Then 5 μl 10% NP-40 was added, and the lysate was vortexed for 10 s before centrifugation at 3,000 rpm for 10 min at 4°C. The supernatant containing the cytoplasmic fraction was collected and treated with 1 ml of TriReagent (T9424; Sigma-Aldrich). The remaining pellet containing the nuclear fraction was treated with 1 ml TriReagent as well. RNA was extracted as per the manufacturer’s orders. 106 cells were used per genotype per time point. cDNA was prepared using the High-Capacity cDNA Reverse Transcription Kit (4368814; Thermo Fisher Scientific), and AIRE and HPRT mRNA were detected by real-time qPCR using the following TaqMan assays: AIRE, Mm00477461_m1; and HPRT, Mm00446968. mRNA decay was determined by using nonlinear regression curve fitting (one-phase decay) of relative mRNA abundance based on ΔΔCt of the relevant time point and its 0 h time point in each cellular fraction separately.
Isolation and analysis of colonic epithelial cells
Colons were harvested from aged mice (14–18 wk) and placed in cold 1× PBS on ice. Colons were cleaned of feces by flushing with cold 1× PBS. The colon was then cut open longitudinally to expose the epithelial layer and placed in 20 ml 2 mM EDTA at 37°C with vigorous shaking for 30 min. The supernatant was then transferred to a clean tube, and cells were pelleted at 300 g for 10 min. Cell pellets were then resuspended in 4 ml of 1 mg/ml Collagenase-Dispase cocktail (269638; Roche), 2% FBS, and 10 µg/ml DNase in RPMI for 10 min with occasional agitation. Single-cell suspensions were washed with MACS buffer followed by centrifugation at 340 g for 5 min at 4°C. Cells were then stained with specific surface antibodies: EpCAM APC-Cy7 (118218; Biolegend) and CD45 PE-Cy7 (103114; Biolegend). For intracellular staining, cells were fixed and permeabilized using BD Cytifix/Cytoperm (554714; BD Biosciences), then stained with anti-Chga (ab15160; Abcam) followed by secondary stain with goat anti-rabbit IgG conjugated to AF555 (ab150078; Abcam). DAPI (D9542; Sigma-Aldrich) was added before acquisition to visualize nuclei. Cells were resuspended in 20–30 µl MACS buffer, acquired on an ImageStreamX Mark II flow cytometer (for details, see above), and analyzed using IDEAS 6.2 software (Amnis Corp.).
Immunofluorescence staining of paraffin-embedded colon sections
Colons from 17–18-wk-old mice were fixed in 4% formaldehyde for 48 h, followed by embedding in paraffin. Dewaxing of 5-µm-thick sections was conducted using xylene, and rehydration was achieved in graded ethanol baths. Antigen retrieval was performed by boiling slides for 20 min in 10 mM sodium citrate buffer, pH 6.0. Nonspecific binding sites were blocked in blocking buffer (Tris-buffered saline, pH 7.4, 10% goat serum, and 0.2% Triton X-100) for 30 min at room temperature. Sections were then incubated with anti-Chga (ab15160; Abcam) diluted in blocking buffer overnight at 4°C. Slides were washed twice with Tris-buffered saline with Tween-20 before incubation with goat anti-rabbit IgG AF488 secondary antibody (111–545-003; Jackson ImmunoResearch) and DAPI in blocking buffer. Stained sections were visualized on a Nikon-Ti-E inverted fluorescence microscope with a 20× objective using NIS-Elements software (Nikon). Image analysis was performed using ImageJ software.
Body composition
Body composition of conscious, unmedicated mice was assessed using the minispec Live Mice Analyzer (LF50; Bruker) per the manufacturer’s instructions.
Preparation of cell and tissue lysates
Single-cell suspensions from two or three thymi following Percoll enrichment for TECs (see above) were further enriched for TECs by staining for EpCAM-APC (118214; Biolegend), followed by positive selection using anti-APC microbeads (130–090-855; Miltenyi Biotec). The enriched cells were washed in MACS buffer (340 g, 5 min, 4°C), and the pellet was lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-Cl, pH 8.0, 5 mM EDTA, 1% IGEPAL CA-360, 0.5% sodium deoxycholate, and 0.1% SDS) complemented with complete proteinase inhibitors (539134; Calbiochem).
Organs were surgically removed from NSG mice, and 100 mg of the tissue was lysed in 1 ml lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1% IGEPAL CA-360) supplemented with complete protease inhibitors and were processed with a tissue homogenizer. Homogeneous lysates were spun at 12,000 g for 20 min at 4°C. The supernatant was collected and supplemented with 4× sample buffer and boiled for 5 min at 96°C.
SDS-PAGE and immunoblot analysis
Proteins were resolved on a 10% SDS-PAGE and were electro-transferred onto nitrocellulose membranes. Membranes of samples derived from thymus were treated with Pierce Western Blot Signal Enhancer (21050; Thermo Fisher Scientific) before nonspecific binding blockade, which was performed in a solution of 5% milk in 1× PBS supplemented with 0.05% Tween-20. All membranes were incubated overnight with antibodies/serum. Signals were detected with an EZ-ECL Kit (Biological Industries) or Immobilion Crescendo Western HRP Substrate (WBLUR0500; Millipore) and were captured by the ImageQuant TM-RT ECL image analysis system (GE Healthcare). Quantification of WBs was done using Image Lab Software 6.1 (BioRad). The following antibodies were used: polyclonal anti-rat AIRE SAND (a kind gift from Diane Mathis), anti-GAPDH (MAB374; Millipore), anti-H2B (ab45695; Abcam), horseradish peroxidase–conjugated monoclonal antibody to rat IgG light chain (112–035-175; Jackson ImmunoResearch), horseradish peroxidase–conjugated goat polyclonal antibody to mouse (115–035-003; Jackson ImmunoResearch), and horseradish peroxidase–conjugated monoclonal antibody to rabbit IgG (111–035-003; Jackson ImmunoResearch).
Analysis of autoantibodies
For the analysis of autoantibodies, serum at a 1:200 dilution in 5% skim milk was used to probe tissue lysates from NSG mice by slot-blot analysis (see above). 1 mg of protein was used per slot-blot analysis.
Radiobinding assay
Human sera were analyzed for antibodies against IFN-ω, IL-22, IL-17F, NACHT leucine-rich-repeat protein 5 (NALP5), 21-hydroxylase (21OH), side-chain cleavage enzyme (SCC), 17-hydroxylase (17OH), aromatic l-amino acid decarboxylase (AADC), and tryptophan hydroxylase (TPH; Alimohammadi et al., 2008; Husebye et al., 1997; Oftedal et al., 2008; Söderbergh et al., 1996; Winqvist et al., 1993, 1992) using in vitro transcribed and translated radiolabeled proteins essentially as described by Ekwall et al. (1998). Briefly, the proteins of interest were expressed by in vitro transcription/translation using the TnT Quick Coupled Transcription/Translation System (Promega) with 35S methionine. The amount of radiolabeled protein was determined by a Tri Carb 4910TR liquid scintillation analyzer (Perkin Elmer) by adding 2 μl of the protein to a filter paper, washing three times with wash buffer (20 mM Tris-HCl, 150 mM NaCl, 0.1% BSA, and 0.15% Tween) using a vacuum manifold, and adding 1 ml Ultima Gold MV (Perkin Elmer). 2 × 104 cpm radiolabeled protein was added to human sera (1:5 dilution) in a total of 50 µl (diluted in wash buffer), and incubated overnight at 4°C. MultiScreen-BV Filter Plates, 1.2 µm (Merck), were prepared by blocking the plates for 2 h with 1% BSA in wash buffer, and by rinsing the wells twice with wash buffer supplemented with 0.05% Tween. The plates were stored at 4°C until use. The serum:protein mix was transferred to MultiScreen-BV Filter Plates, 1.2 µm (Merck), added to 50 μl Protein A Sepharose Fast Flow (Cytiva Life Sciences), and incubated for 45 min at 4°C with shaking. The filter plate was subsequently washed three times with wash buffer using a vacuum manifold. After the filter plate was dried, 20 μl scintillation fluid (Micorscint O; Perkin Elmer) was added to each well, and the plate analyzed on a microplate scintillation and luminescence counter, Top Count (Packard). A positive (index 1,000) and negative (index 0) control serum was used to calculate the autoantibody indexes, and the cutoff for positivity calculated by an average of at least 150 healthy controls + 3 SD.
Analysis of proteasomal and protease-mediated degradation
For assessment of proteasomal or protease-mediated degradation following EpCAM enrichment (see above), cells from four mice per genotype were pooled, and an equal number of cells was divided between treatment groups. Cells were treated with RPMI medium (1% L-glutamine, 1% pen-strep, and 10% FBS) with a final concentration of the proteasome inhibitor 10 mM MG132 (474790; Calbiochem) in DMSO (D2438; Sigma-Aldrich), or with 1:400 Protease Inhibitor Cocktail III (539134; Calbiochem) or DMSO alone. Cells were then incubated at 37°C with 5% CO2 for 2.5 h (protease inhibition) or 3 h (proteasome inhibition) before harvesting, lysing, and further preparation for WB (as described above).
Histology and histopathology
Organs and blood were harvested postmortem from mice of the specified strain and age. Blood was collected by cardiac puncture and allowed to coagulate for 30 min before centrifugation at 1,500 g for 15 min and serum collection. All organs were fixed in 4% formaldehyde for 48 h, washed in 70% ethanol, and embedded in paraffin. Organs were sectioned and stained for H&E by the histology laboratory at the Weizmann Institute of Science. Histopathology, including analysis and scoring of immune infiltrates in various peripheral tissues, was performed under the supervision of a certified animal pathologist. Briefly, scoring was assessed according to size and frequency of immune infiltrates, with scanning of three slices from each animal. Score 0 marked animals in which no infiltrates were found, while score 3 marked animals in which significant destruction of the organ was observed. Stained sections were visualized on an Eclipse Nikon-Ni upright light microscope with a 4× (eye, prostate, and salivary gland) or 10× (liver, pancreas, lung, and colon) objective using NIS-Elements software (Nikon).
5HT ELISA
Serum collected from aged mice (see above) was used to assess 5HT levels in the blood using a commercial ELISA kit (ab133053; Abcam) per the manufacturer’s instructions with serum used at a 1:10 dilution.
ATACseq
Assessment of chromatin accessibility in mTEChi was conducted using ATACseq (Buenrostro et al., 2015), with modifications (Lara-Astiaso et al., 2014). Briefly, mTEChi were sorted into 100 µl of MACS buffer and pelleted by centrifugation for 20 min at 500 g at 4°C with low acceleration and brake settings. Cell pellets were resuspended in 50 µl of lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630), and nuclei were pelleted by centrifugation for 20 min at 500 g, 4°C, with low acceleration and brake settings. The supernatant was discarded, and nuclei were resuspended in 25 µl reaction buffer containing 2 µl of Tn5 transposase and 12.5 µl of TD buffer (Nextera Sample preparation kit; Illumina). The reaction was incubated at 37°C for 1 h. Then, 5 µl of clean-up buffer (900 mM NaCl and 300 mM EDTA), 2 µl of 5% SDS, and 2 µl of Proteinase K (AM2548; Invitrogen) were added and incubated for 30 min at 40°C. Tagmentated DNA was isolated using 2× solid phase reversible immobilization (SPRI) bead clean-up (A63881; AMPure XP; Beckman Coulter). For library amplification, two sequential nine-cycle PCR runs were performed in order to enrich small tagmentated DNA fragments. We used 2 µl of indexing primers included in the Nextera Index kit and KAPA HiFi HotStart ready mix (KK2601; Kapa Biosystems). After the first PCR, the libraries were selected for small fragments (<600 bp) using SPRI clean-up. Then a second PCR was performed with the same conditions in order to obtain the final library. Samples were sequences on a NextSeq500 platform.
For ATACseq analysis, paired-end raw reads were trimmed using cutadapt (Martin, 2011; with the parameter -q 25 –m 30), and mapped to genome mm10 (GENCODE) using bowtie2 (Langmead and Salzberg, 2012; with the parameters --local). Following alignment, mitochondrial genes were removed from the analysis, and duplicated reads were removed using picard-tools. Nucleosome-free fragments at the length <120 bp were selected from the remaining unique reads, and broad peaks were called using MACS2 (Zhang et al., 2008; with the parameters --bw 120 -B -f BAMPE --SPMR --B --shift -50 --extsize 100 -keep-dup all -q 0.05). Quantification of reads on the Aire promoter (chr10:78043482-78043774) and Aire enhancer (chr10:78046193-78046998) regions was done using DiffBind (DESeq method; Stark and Brown, 2011). For visualization of ATACseq alignments, coverage files were converted to tiled data files using the IGVTools (Robinson et al., 2011) package and displayed using IGV 2.4.16, normalized by [1,000,000/(totalReadCount)], and scaled to the data range.
ChIPseq
1.5 × 105 sorted mTEChi from F1.Aire+/C313Y (see above), B6.Aire+/C442G mice, or their WT littermates were used for each ChIPseq sample, using adapted published protocols (Bansal et al., 2017; Blecher-Gonen et al., 2013). Briefly, before sorting, EpCAM-enriched cells were cross-linked with 1% formaldehyde (28906; Thermo Fisher Scientific) for 10 min, then quenched with 2.5 M glycine for 5 min. mTEChi were sorted into 10% FBS and then pelleted for 15 min at 1,500 g with minimal brake. Cells were then resuspended in hypotonic buffer (12 mM Tris-HCl, pH 8, and 6 mM EDTA, pH 8.0, in 0.1× PBS) supplemented with protease inhibitor cocktail (04693124001; cOmplete Tablets; Roche) and pelleted again for 15 min at 1,500 g with minimal brake. The cell pellet was then frozen at −80°C until further processing. Then the cells were lysed on ice in RIPA buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, pH 8, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, and 0.1% sodium deoxycholate) supplemented with complete protease inhibitor cocktail for 10 min. Chromatin was sheared using a Biorupter300 (Diagenode) at high intensity for 40 cycles (30 s on, 30 s off). The sheared material was cleared by a 10-min centrifugation at 13,000 rpm at 4°C. Prior to ChIP, 5% of the sheared material from each sample was taken to serve as the input control. Then the cleared material was immunoprecipitated overnight at 4°C with magnetic Protein-G beads (Dynabeads; Life Technologies) conjugated to 10 μg anti-AIRE (5H12 14–5934-82; eBioscience) followed by extensive washing of the beads with ice-cold RIPA, high-salt RIPA (10 mM Tris-HCl, pH 8, 1 mM EDTA, pH 8, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, and 0.1% sodium deoxycholate), LiCl (10 mM Tris-HCl, pH 8, 1 mM EDTA, pH 8, 250 mM LiCl, 0.5% NP-40, and 0.5% sodium deoxycholate) and TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA, pH 8). Chromatin was eluted from the beads with elution buffer (10 mM Tris-HCl, pH 8, 5 mM EDTA, pH 8, 300 mM NaCl, and 0.1% SDS) and along with the input controls was treated with 1 µg DNase-free RNase (11119915001; Roche) for 30 min at 37°C and with Proteinase K (25530–049; Invitrogen) and glycogen (10901393001; Roche) for 2 h at 37°C followed by reverse cross-linking by leaving the plate at 65°C overnight. DNA from reverse cross-linked material was purified with SPRI beads (SPRIselect beads; Beckman Coulter), and libraries were prepared by sequential steps of end repair, A-base addition, adaptor ligation, and PCR amplification (14 cycles; Blecher-Gonen et al., 2013). ChIPseq libraries were size-selected for 200–600-bp fragments with SPRI beads. Equivalent amounts of barcoded libraries were pooled and sequenced on a NextSeq 500 (Illumina).
For analysis of paired-end ChIPseq libraries, reads were preprocessed with cutadapt (Martin, 2011) to remove adapters and low-quality bases (parameters: --times 2 -q 20 -m 25) and mapped to mm10 genome (GENCODE) using bowtie2 (Langmead and Salzberg, 2012; parameters: --local, -X 2000). Uniquely mapped reads were extracted using samtools (-F 4, -f 0 × 2, -q 39). Narrow peaks were called using MACS2 (Zhang et al., 2008) compared with background input (whole cell extract [WCE]) samples (parameters: --bw 300 -B -f --SPMR -keep-dup auto -q 0.05 BAMPE --nomodel). Marking and removing reads duplicates was done using Picard (MarkDuplicates), and peaks overlapping ENCODE Blacklist regions were omitted (Amemiya et al., 2019). For visualization of ChIPseq alignments, coverage files were converted to tiled data files using the IGVTools package (Robinson et al., 2011), normalized by [1,000,000/(totalReadCount)], and scaled to data range. Spearman correlation coefficients were computed based on read coverage between samples using “plotCorrelation” using the deepTools2 package (Ramírez et al., 2016). Heatmaps were constructed with deepTools2 in “reference-point” mode around peak summits, with missingDataAsZero parameter, and were clustered using the K-means method in “plotHeatmap.” Read coverage plots were created using ngs.plot (Shen et al., 2014) using merged alignment files of replicates. Quantification of peak regions was done using DiffBind (v3.0.15), normalized based on full library sizes and the DESeq2 method (Stark and Brown, 2011).
Statistical analysis
RNAseq data statistical analysis was described above. Student’s t tests were used for comparison between two groups. ANOVA was used for comparisons between three or more groups with Tukey post hoc comparisons between all groups. Repeated-measures ANOVA was used for comparing weight change. The log-rank (Mantel–Cox) test was used to assess survival. The Kruskal–Wallis nonparametric test with Dunn’s post hoc test or Wilcoxon signed rank test was used to assess the statistical significance of H&E histopathology. All statistical analyses were performed using GraphPad Prism 5.0. P values <0.05 were considered significant.
Online supplemental material
Fig. S1 shows that the heterozygous Aire+/C311Y mutation affects fertility and mTEC cellularity. Fig. S2 shows that the heterozygous Aire+/C313Y mutation, but not Aire+/V303M, impairs ectopic expression of AIRE target genes in mTECs and the thymic T reg cell compartment. Fig. S3 shows that the dominant-negative Aire+/C313Y mutation results in devastating multi-organ autoimmunity. Fig. S4 shows identification of C446G AIRE as a novel dominant mutation located in the PHD2 domain. Fig. S5 shows that dominant and recessive AIRE mutations differentially affect AIRE protein expression, and AIRE directly regulates its own expression. Table S1 lists single-guide RNA and repair oligos used for CRISPR/Cas9 creation of mice strains. Table S2 shows ChIPseq quality control. Table S3 shows the phenotype and autoantibody profile of patients carrying the AIRE C446G mutation. Table S4 shows custom TaqMan genotyping assays.
Supplementary Material
lists single-guide RNA and repair oligos used for CRISPR/Cas9 creation of mice strains.
shows ChIPseq quality control.
shows the phenotype and autoantibody profile of patients carrying the AIRE C446G mutation.
shows custom TaqMan genotyping assays.
Acknowledgments
We thank the Abramson laboratory group for critical reading of the manuscript and for many fruitful discussions. We thank the personnel at the Weizmann Institute of Science animal facility for animal husbandry and the Abramson laboratory genotyping team for their work. We thank the personnel of the Weizmann Institute of Science histology unit for their skilled work in preparation of H&E slides and Dr. Ori Brenner for helping with their classification. We thank Dr. Chamutal Bornstein and Dr. Ronnie Blecher-Gonen for their help with ChIPseq calibrations. We thank Ms. Tali Wiesel from the Weizmann Institute of Science graphics department for her help with illustrations.
Research in the Abramson laboratory is kindly supported by the European Research Council (ERC-2016-CoG-724821), Israel Science Foundation (1796/16), Bill and Marika Glied and Family Fund, Wohl Biology Endowment Fund, Erica Drake Fund, Enoch Foundation, Ruth and Samuel David Gameroff Family Foundation and Lilly Fulop Fund for Multiple Sclerosis Research. U. Haljasorg was funded by the Estonian Research Council (grant PUTJD707). Y. Gruper was supported by the Ariane de Rothschild Women Doctoral Program Fellowship. J. Dobeš was supported by the Dean of Faculty Fellowship for outstanding postdocs of the Weizmann Institute of Science and by the Weizmann Institute of Science–Czech Academy of Sciences Bilateral Fellowship. C. Peligero-Cruz was supported by the Weizmann–La Caixa Fellowship and Dean of Faculty Fellowship for outstanding postdocs of the Weizmann Institute of Science. Research in the Husebye laboratory is kindly supported by the Kristian Gerhard Jebsen Foundation, Norwegian Research Council (262677), and Novonordisk Foundation. E.S. Husebye was also supported by the Weston Visiting Scholar Program at the Weizmann Institute of Science.
Author contributions: Y. Goldfarb, E.S. Husebye, and J. Abramson conceived and designed the project. Y. Goldfarb performed most of the experiments. Y. Kuperman, T. Givony, N. Kadouri, J. Dobeš, C. Peligero-Cruz, I. Zalayat, Y. Gruper, A. Avin, S. Nevo, A. Ulman, and U. Haljasorg aided in performing some experiments and analyses. B.E. Oftedal, E. Bratland, and M.M. Erichsen conducted all patient-related experiments. A. Berger created all Aire domain models. S. Ben-Dor designed all CRISPR mice. G. Damari and R. Haffner-Krausz created all CRISPR strains. B. Dassa performed bioinformatics analysis of ATACseq and ChIPseq data. D. Leshkowitz performed bioinformatics analysis of ChIPseq data. Z. Porat helped with ImageStream analysis. U. Atasoy advised regarding mRNA decay. U. Atasoy, J. Dobeš, N. Kadouri, B. Dassa, S. Ben-Dor, and A. Berger critically evaluated the manuscript. Y. Goldfarb, E.S. Husebye, and J. Abramson wrote the manuscript.
Data availability
RNAseq, ATACseq, and ChIPseq datasets have been deposited in GEO under accession no. GSE151013.
References
- Abbott, J.K., Huoh Y.S., Reynolds P.R., Yu L., Rewers M., Reddy M., Anderson M.S., Hur S., and Gelfand E.W.. 2018. Dominant-negative loss of function arises from a second, more frequent variant within the SAND domain of autoimmune regulator (AIRE). J. Autoimmun. 88:114–120. 10.1016/j.jaut.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimohammadi, M., Björklund P., Hallgren A., Pöntynen N., Szinnai G., Shikama N., Keller M.P., Ekwall O., Kinkel S.A., Husebye E.S., et al. 2008. Autoimmune polyendocrine syndrome type 1 and NALP5, a parathyroid autoantigen. N. Engl. J. Med. 358:1018–1028. 10.1056/NEJMoa0706487 [DOI] [PubMed] [Google Scholar]
- Amemiya, H.M., Kundaje A., and Boyle A.P.. 2019. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 9:9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J., von Boehmer H., Bronson R., Dierich A., Benoist C., and Mathis D.. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298:1395–1401. 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- Aricha, R., Feferman T., Scott H.S., Souroujon M.C., Berrih-Aknin S., and Fuchs S.. 2011. The susceptibility of Aire(-/-) mice to experimental myasthenia gravis involves alterations in regulatory T cells. J. Autoimmun. 36:16–24. 10.1016/j.jaut.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Aschenbrenner, K., D’Cruz L.M., Vollmann E.H., Hinterberger M., Emmerich J., Swee L.K., Rolink A., and Klein L.. 2007. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8:351–358. 10.1038/ni1444 [DOI] [PubMed] [Google Scholar]
- Asou, H., Gombart A.F., Takeuchi S., Tanaka H., Tanioka M., Matsui H., Kimura A., Inaba T., and Koeffler H.P.. 2003. Establishment of the acute myeloid leukemia cell line Kasumi-6 from a patient with a dominant-negative mutation in the DNA-binding region of the C/EBPalpha gene. Genes Chromosomes Cancer. 36:167–174. 10.1002/gcc.10161 [DOI] [PubMed] [Google Scholar]
- Bansal, K., Yoshida H., Benoist C., and Mathis D.. 2017. The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol. 18:263–273. 10.1038/ni.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barndt, R.J., Dai M., and Zhuang Y.. 2000. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 20:6677–6685. 10.1128/MCB.20.18.6677-6685.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björses, P., Halonen M., Palvimo J.J., Kolmer M., Aaltonen J., Ellonen P., Perheentupa J., Ulmanen I., and Peltonen L.. 2000. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am. J. Hum. Genet. 66:378–392. 10.1086/302765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecher-Gonen, R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., and Amit I.. 2013. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8:539–554. 10.1038/nprot.2013.023 [DOI] [PubMed] [Google Scholar]
- Bottomley, M.J., Stier G., Pennacchini D., Legube G., Simon B., Akhtar A., Sattler M., and Musco G.. 2005. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. J. Biol. Chem. 280:11505–11512. 10.1074/jbc.M413959200 [DOI] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq method. Curr. Protoc. Mol. Biol. 109:29.1–: 9.. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldenhoven, E., van Dijk T.B., Solari R., Armstrong J., Raaijmakers J.A.M., Lammers J.W.J., Koenderman L., and de Groot R.P.. 1996. STAT3β, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 271:13221–13227. 10.1074/jbc.271.22.13221 [DOI] [PubMed] [Google Scholar]
- Cetani, F., Barbesino G., Borsari S., Pardi E., Cianferotti L., Pinchera A., and Marcocci C.. 2001. A novel mutation of the autoimmune regulator gene in an Italian kindred with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, acting in a dominant fashion and strongly cosegregating with hypothyroid autoimmune thyroiditis. J. Clin. Endocrinol. Metab. 86:4747–4752. 10.1210/jcem.86.10.7884 [DOI] [PubMed] [Google Scholar]
- Cowan, J.E., Baik S., McCarthy N.I., Parnell S.M., White A.J., Jenkinson W.E., and Anderson G.. 2018. Aire controls the recirculation of murine Foxp3+ regulatory T-cells back to the thymus. Eur. J. Immunol. 48:844–854. 10.1002/eji.201747375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Siervi, A., De Luca P., Byun J.S., Di L.J., Fufa T., Haggerty C.M., Vazquez E., Moiola C., Longo D.L., and Gardner K.. 2010. Transcriptional autoregulation by BRCA1. Cancer Res. 70:532–542. 10.1158/0008-5472.CAN-09-1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre, E., Ivanauskiene K., Spirkoski J., Shah A., Vekterud K., Moskaug J.Ø., Bøe S.O., Wong L.H., Küntziger T., and Collas P.. 2017. PML protein organizes heterochromatin domains where it regulates histone H3.3 deposition by ATRX/DAXX. Genome Res. 27:913–921. 10.1101/gr.215830.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobeš, J., Neuwirth A., Dobešová M., Vobořil M., Balounová J., Ballek O., Lebl J., Meloni A., Krohn K., Kluger N., et al. 2015. Gastrointestinal Autoimmunity Associated With Loss of Central Tolerance to Enteric α-Defensins. Gastroenterology. 149:139–150. 10.1053/j.gastro.2015.05.009 [DOI] [PubMed] [Google Scholar]
- Ekwall, O., Hedstrand H., Grimelius L., Haavik J., Perheentupa J., Gustafsson J., Husebye E., Kämpe O., and Rorsman F.. 1998. Identification of tryptophan hydroxylase as an intestinal autoantigen. Lancet. 352:279–283. 10.1016/S0140-6736(97)11050-9 [DOI] [PubMed] [Google Scholar]
- Eriksson, D., Røyrvik E.C., Aranda-Guillén M., Berger A.H., Landegren N., Artaza H., Hallgren Å., Grytaas M.A., Ström S., Bratland E., et al. Swedish Addison Registry Study Group . 2021. GWAS for autoimmune Addison’s disease identifies multiple risk loci and highlights AIRE in disease susceptibility. Nat. Commun. 12:959. 10.1038/s41467-021-21015-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw, C.H., Dellaire G., and Bazett-Jones D.P.. 2004. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J. Biol. Chem. 279:9577–9585. 10.1074/jbc.M312580200 [DOI] [PubMed] [Google Scholar]
- Ferguson, B.J., Alexander C., Rossi S.W., Liiv I., Rebane A., Worth C.L., Wong J., Laan M., Peterson P., Jenkinson E.J., et al. 2008. AIRE’s CARD revealed, a new structure for central tolerance provokes transcriptional plasticity. J. Biol. Chem. 283:1723–1731. 10.1074/jbc.M707211200 [DOI] [PubMed] [Google Scholar]
- Finnish-German APECED Consortium . 1997. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17:399–403. 10.1038/ng1297-399 [DOI] [PubMed] [Google Scholar]
- Gaetani, M., Matafora V., Saare M., Spiliotopoulos D., Mollica L., Quilici G., Chignola F., Mannella V., Zucchelli C., Peterson P., et al. 2012. AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome. Nucleic Acids Res. 40:11756–11768. 10.1093/nar/gks933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, J.M., Devoss J.J., Friedman R.S., Wong D.J., Tan Y.X., Zhou X., Johannes K.P., Su M.A., Chang H.Y., Krummel M.F., and Anderson M.S.. 2008. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 321:843–847. 10.1126/science.1159407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, T.J., Ramu C., Gemünd C., and Aasland R.. 1998. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem. Sci. 23:242–244. 10.1016/S0968-0004(98)01231-6 [DOI] [PubMed] [Google Scholar]
- Grasberger, H., Ringkananont U., Lefrancois P., Abramowicz M., Vassart G., and Refetoff S.. 2005. Thyroid transcription factor 1 rescues PAX8/p300 synergism impaired by a natural PAX8 paired domain mutation with dominant negative activity. Mol. Endocrinol. 19:1779–1791. 10.1210/me.2004-0426 [DOI] [PubMed] [Google Scholar]
- Gunawardene, A.R., Corfe B.M., and Staton C.A.. 2011. Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int. J. Exp. Pathol. 92:219–231. 10.1111/j.1365-2613.2011.00767.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haljasorg, U., Bichele R., Saare M., Guha M., Maslovskaja J., Kõnd K., Remm A., Pihlap M., Tomson L., Kisand K., et al. 2015. A highly conserved NF-κB-responsive enhancer is critical for thymic expression of Aire in mice. Eur. J. Immunol. 45:3246–3256. 10.1002/eji.201545928 [DOI] [PubMed] [Google Scholar]
- Halonen, M., Kangas H., Rüppell T., Ilmarinen T., Ollila J., Kolmer M., Vihinen M., Palvimo J., Saarela J., Ulmanen I., and Eskelin P.. 2004. APECED-causing mutations in AIRE reveal the functional domains of the protein. Hum. Mutat. 23:245–257. 10.1002/humu.20003 [DOI] [PubMed] [Google Scholar]
- Herzig, Y., Nevo S., Bornstein C., Brezis M.R., Ben-Hur S., Shkedy A., Eisenberg-Bord M., Levi B., Delacher M., Goldfarb Y., et al. 2017. Transcriptional programs that control expression of the autoimmune regulator gene Aire. Nat. Immunol. 18:161–172. 10.1038/ni.3638 [DOI] [PubMed] [Google Scholar]
- Hubert, F.-X., Kinkel S.A., Crewther P.E., Cannon P.Z.F., Webster K.E., Link M., Uibo R., O’Bryan M.K., Meager A., Forehan S.P., et al. 2009. Aire-deficient C57BL/6 mice mimicking the common human 13-base pair deletion mutation present with only a mild autoimmune phenotype. J. Immunol. 182:3902–3918. 10.4049/jimmunol.0802124 [DOI] [PubMed] [Google Scholar]
- Huoh, Y.S., Wu B., Park S., Yang D., Bansal K., Greenwald E., Wong W.P., Mathis D., and Hur S.. 2020. Dual functions of Aire CARD multimerization in the transcriptional regulation of T cell tolerance. Nat. Commun. 11:1625. 10.1038/s41467-020-15448-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye, E.S., Gebre-Medhin G., Tuomi T., Perheentupa J., Landin-Olsson M., Gustafsson J., Rorsman F., and Kämpe O.. 1997. Autoantibodies against aromatic L-amino acid decarboxylase in autoimmune polyendocrine syndrome type I. J. Clin. Endocrinol. Metab. 82:147–150. 10.1210/jc.82.1.147 [DOI] [PubMed] [Google Scholar]
- Husebye, E.S., Anderson M.S., and Kämpe O.. 2018. Autoimmune polyendocrine syndromes. N. Engl. J. Med. 378:1132–1141. 10.1056/NEJMra1713301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacenko, O., LuValle P., Solum K., and Olsen B.R.. 1993. A dominant negative mutation in the alpha 1 (X) collagen gene produces spondylometaphyseal defects in mice. Prog. Clin. Biol. Res. 383B:427–436. [PubMed] [Google Scholar]
- Jaitin, D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A., and Amit I.. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 343:776–779. 10.1126/science.1247651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti, S., Warren B.D., McGinnis L.K., Kinsey W.H., Petroff B.K., and Petroff M.G.. 2012. The autoimmune regulator prevents premature reproductive senescence in female mice. Biol. Reprod. 86:110. 10.1095/biolreprod.111.097501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Anderson M.S., Bronson R., Mathis D., and Benoist C.. 2005. Modifier loci condition autoimmunity provoked by Aire deficiency. J. Exp. Med. 202:805–815. 10.1084/jem.20050693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanguy, E., Lamhamedi-Cherradi S., Lammas D., Dorman S.E., Fondanèche M.C., Dupuis S., Döffinger R., Altare F., Girdlestone J., Emile J.F., et al. 1999. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat. Genet. 21:370–378. 10.1038/7701 [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani, P., Beitel L.K., Trifiro M., Kaufman M., Rennie P., Sheppard P., Matusik R., and Pinsky L.. 1993. Substitution of valine-865 by methionine or leucine in the human androgen receptor causes complete or partial androgen insensitivity, respectively with distinct androgen receptor phenotypes. Mol. Endocrinol. 7:37–46. 10.1210/mend.7.1.8446106 [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., Mezulis S., Yates C.M., Wass M.N., and Sternberg M.J.E.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10:845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa, S.M., and Vingron M.. 2008. Transcriptional autoregulatory loops are highly conserved in vertebrate evolution. PLoS One. 3:e3210. 10.1371/journal.pone.0003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, A.S., Kuo A.J., Park S.Y., Cheung P., Abramson J., Bua D., Carney D., Shoelson S.E., Gozani O., Kingston R.E., et al. 2008. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl. Acad. Sci. USA. 105:15878–15883. 10.1073/pnas.0808470105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, A.S., Miller E.L., Buenrostro J.D., Moskowitz D.M., Wang J., Greenleaf W.J., Chang H.Y., and Crabtree G.R.. 2018. Rapid chromatin repression by Aire provides precise control of immune tolerance. Nat. Immunol. 19:162–172. 10.1038/s41590-017-0032-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohen, R., Barlev J., Hornung G., Stelzer G., Feldmesser E., Kogan K., Safran M., and Leshkowitz D.. 2019. UTAP: User-friendly Transcriptome Analysis Pipeline. BMC Bioinformatics. 20:154. 10.1186/s12859-019-2728-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P.G., Laloraya M., Wang C.Y., Ruan Q.G., Davoodi-Semiromi A., Kao K.J., and She J.X.. 2001. The autoimmune regulator (AIRE) is a DNA-binding protein. J. Biol. Chem. 276:41357–41364. 10.1074/jbc.M104898200 [DOI] [PubMed] [Google Scholar]
- LaFlam, T.N., Seumois G., Miller C.N., Lwin W., Fasano K.J., Waterfield M., Proekt I., Vijayanand P., and Anderson M.S.. 2015. Identification of a novel cis-regulatory element essential for immune tolerance. J. Exp. Med. 212:1993–2002. 10.1084/jem.20151069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, M., Jegou T., Chung I., Richter K., Münch S., Udvarhelyi A., Cremer C., Hemmerich P., Engelhardt J., Hell S.W., and Rippe K.. 2010. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 123:392–400. 10.1242/jcs.053496 [DOI] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso, D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S., et al. 2014. Immunogenetics. Chromatin state dynamics during blood formation. Science. 345:943–949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani, J.J., Depetris D., Usson Y., Metzler-Guillemain C., Mignon-Ravix C., Mitchell M.J., Megarbane A., Sarda P., Sirma H., Moncla A., et al. 2006. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J. Cell Sci. 119:2518–2531. 10.1242/jcs.02965 [DOI] [PubMed] [Google Scholar]
- Mann, D.A., and Oakley F.. 2013. Serotonin paracrine signaling in tissue fibrosis. Biochim. Biophys. Acta. 1832:905–910. 10.1016/j.bbadis.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mathis, D., and Benoist C.. 2009. Aire. Annu. Rev. Immunol. 27:287–312. 10.1146/annurev.immunol.25.022106.141532 [DOI] [PubMed] [Google Scholar]
- Moran, G.W., Leslie F.C., Levison S.E., Worthington J., and McLaughlin J.T.. 2008. Enteroendocrine cells: neglected players in gastrointestinal disorders? Therap. Adv. Gastroenterol. 1:51–60. 10.1177/1756283X08093943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine, K., Peterson P., Scott H.S., Kudoh J., Minoshima S., Heino M., Krohn K.J.E., Lalioti M.D., Mullis P.E., Antonarakis S.E., et al. 1997. Positional cloning of the APECED gene. Nat. Genet. 17:393–398. 10.1038/ng1297-393 [DOI] [PubMed] [Google Scholar]
- Oftedal, B.E., Wolff A.S., Bratland E., Kämpe O., Perheentupa J., Myhre A.G., Meager A., Purushothaman R., Ten S., and Husebye E.S.. 2008. Radioimmunoassay for autoantibodies against interferon omega; its use in the diagnosis of autoimmune polyendocrine syndrome type I. Clin. Immunol. 129:163–169. 10.1016/j.clim.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Oftedal, B.E., Hellesen A., Erichsen M.M., Bratland E., Vardi A., Perheentupa J., Kemp E.H., Fiskerstrand T., Viken M.K., Weetman A.P., et al. 2015. Dominant Mutations in the Autoimmune Regulator AIRE Are Associated with Common Organ-Specific Autoimmune Diseases. Immunity. 42:1185–1196. 10.1016/j.immuni.2015.04.021 [DOI] [PubMed] [Google Scholar]
- Org, T., Chignola F., Hetényi C., Gaetani M., Rebane A., Liiv I., Maran U., Mollica L., Bottomley M.J., Musco G., and Peterson P.. 2008. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9:370–376. 10.1038/embor.2008.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornatsky, O.I., Andreucci J.J., and McDermott J.C.. 1997. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272:33271–33278. 10.1074/jbc.272.52.33271 [DOI] [PubMed] [Google Scholar]
- Ossart, J., Moreau A., Autrusseau E., Ménoret S., Martin J.C., Besnard M., Ouisse L.-H., Tesson L., Flippe L., Kisand K., et al. 2018. Breakdown of Immune Tolerance in AIRE-Deficient Rats Induces a Severe Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy-like Autoimmune Disease. J. Immunol. 201:874–887. 10.4049/jimmunol.1701318 [DOI] [PubMed] [Google Scholar]
- Pan, G., Li J., Zhou Y., Zheng H., and Pei D.. 2006. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 20:1730–1732. 10.1096/fj.05-5543fje [DOI] [PubMed] [Google Scholar]
- Posovszky, C., Lahr G., von Schnurbein J., Buderus S., Findeisen A., Schröder C., Schütz C., Schulz A., Debatin K.M., Wabitsch M., and Barth T.F.. 2012. Loss of enteroendocrine cells in autoimmune-polyendocrine-candidiasis-ectodermal-dystrophy (APECED) syndrome with gastrointestinal dysfunction. J. Clin. Endocrinol. Metab. 97:E292–E300. 10.1210/jc.2011-2044 [DOI] [PubMed] [Google Scholar]
- Ramírez, F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44(W1):W160-5. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, C., Bukrinsky A., and Peltonen L.. 2002a. Systematic mutagenesis of the functional domains of AIRE reveals their role in intracellular targeting. Hum. Mol. Genet. 11:3299–3308. 10.1093/hmg/11.26.3299 [DOI] [PubMed] [Google Scholar]
- Ramsey, C., Winqvist O., Puhakka L., Halonen M., Moro A., Kämpe O., Eskelin P., Pelto-Huikko M., and Peltonen L.. 2002b. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum. Mol. Genet. 11:397–409. 10.1093/hmg/11.4.397 [DOI] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld, N., Elowitz M.B., and Alon U.. 2002. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323:785–793. 10.1016/S0022-2836(02)00994-4 [DOI] [PubMed] [Google Scholar]
- Sansom, S.N., Shikama-Dorn N., Zhanybekova S., Nusspaumer G., Macaulay I.C., Deadman M.E., Heger A., Ponting C.P., and Holländer G.A.. 2014. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24:1918–1931. 10.1101/gr.171645.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, P.A., Leventhal D.S., and Malchow S.. 2014. Shaping the repertoire of tumor-infiltrating effector and regulatory T cells. Immunol. Rev. 259:245–258. 10.1111/imr.12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L., Shao N., Liu X., and Nestler E.. 2014. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 15:284. 10.1186/1471-2164-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.M., Frederiksen J.K., Zacharias D.A., Chan F.K., Johnson M., Lynch D., Tsien R.Y., and Lenardo M.J.. 2000. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 288:2354–2357. 10.1126/science.288.5475.2354 [DOI] [PubMed] [Google Scholar]
- Söderbergh, A., Winqvist O., Norheim I., Rorsman F., Husebye E.S., Dolva O., Karlsson F.A., and Kämpe O.. 1996. Adrenal autoantibodies and organ-specific autoimmunity in patients with Addison’s disease. Clin. Endocrinol. (Oxf.). 45:453–460. 10.1046/j.1365-2265.1996.8040813.x [DOI] [PubMed] [Google Scholar]
- Stark, R., and Brown G.. 2011. DiffBind: differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/html/DiffBind.html (accessed February 3, 2021)
- St-Pierre, C., Trofimov A., Brochu S., Lemieux S., and Perreault C.. 2015. Differential Features of AIRE-Induced and AIRE-Independent Promiscuous Gene Expression in Thymic Epithelial Cells. J. Immunol. 195:498–506. 10.4049/jimmunol.1500558 [DOI] [PubMed] [Google Scholar]
- Su, M.A., Giang K., Zumer K., Jiang H., Oven I., Rinn J.L., Devoss J.J., Johannes K.P.A., Lu W., Gardner J., et al. 2008. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest. 118:1712–1726. 10.1172/JCI34523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winqvist, O., Karlsson F.A., and Kämpe O.. 1992. 21-Hydroxylase, a major autoantigen in idiopathic Addison’s disease. Lancet. 339:1559–1562. 10.1016/0140-6736(92)91829-W [DOI] [PubMed] [Google Scholar]
- Winqvist, O., Gustafsson J., Rorsman F., Karlsson F.A., and Kämpe O.. 1993. Two different cytochrome P450 enzymes are the adrenal antigens in autoimmune polyendocrine syndrome type I and Addison’s disease. J. Clin. Invest. 92:2377–2385. 10.1172/JCI116843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, A.S.B., Erichsen M.M., Meager A., Magitta N.F., Myhre A.G., Bollerslev J., Fougner K.J., Lima K., Knappskog P.M., and Husebye E.S.. 2007. Autoimmune polyendocrine syndrome type 1 in Norway: phenotypic variation, autoantibodies, and novel mutations in the autoimmune regulator gene. J. Clin. Endocrinol. Metab. 92:595–603. 10.1210/jc.2006-1873 [DOI] [PubMed] [Google Scholar]
- Yang, S., Bansal K., Lopes J., Benoist C., and Mathis D.. 2013. Aire’s plant homeodomain(PHD)-2 is critical for induction of immunological tolerance. Proc. Natl. Acad. Sci. USA. 110:1833–1838. 10.1073/pnas.1222023110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Fujikado N., Kolodin D., Benoist C., and Mathis D.. 2015. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 348:589–594. 10.1126/science.aaa7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists single-guide RNA and repair oligos used for CRISPR/Cas9 creation of mice strains.
shows ChIPseq quality control.
shows the phenotype and autoantibody profile of patients carrying the AIRE C446G mutation.
shows custom TaqMan genotyping assays.
Data Availability Statement
RNAseq, ATACseq, and ChIPseq datasets have been deposited in GEO under accession no. GSE151013.