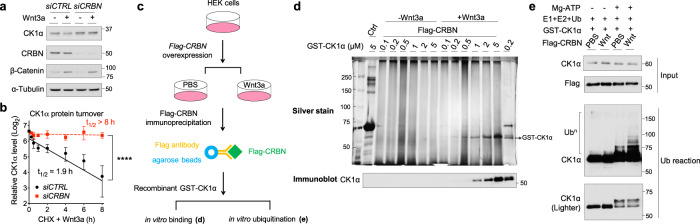
Fig. 2. Wnt-induced degradation of CK1α requires CRBN, the substrate receptor of the CRL4CRBN E3 ubiquitin ligase complex.
a HEK cells were transfected with the indicated smart-pool siRNA and subsequently treated with PBS or Wnt3a for 24 h. Extracts of these cells were evaluated by immunoblotting. A representative immunoblot (n = 5 independent experiments) is shown. b HEK cells were transfected with the indicated smart-pool siRNA and co-treated with Wnt3a and cycloheximide for the indicated time. Extracts of these cells were evaluated by immunoblotting. CK1α levels from immunoblots were quantitated and normalized to HSP90 levels (mean ± SEM, n = 3 independent experiments). Asterisks indicate statistical significance (two-way ANOVA analysis, ****p value < 0.0001). c A schematic outlining experimental details of the in vitro binding and ubiquitination assays shown respectively in panels d and e. d HEK cells were transfected with a plasmid encoding Flag-CRBN and subsequently treated with PBS or Wnt3a for 4 h. Flag-CRBN immunoprecipitated from extracts of these cells were incubated with the indicated amounts of recombinant GST-CK1α at 4 °C for 1 h. Flag-CRBN beads were re-isolated, washed, and CK1α bound was eluted using sample buffer. These immunoprecipitates were subjected to SDS-PAGE, followed by silver staining (top panel) or immunoblotting (bottom panel). IgG beads serve as a control for Flag-CRBN beads (left lane) and recombinant GST-CK1α a loading control (right lane). A representative gel image and immunoblot (n = 3 independent experiments) is shown. e HEK cells were transfected with a plasmid encoding Flag-CRBN and subsequently treated with PBS or Wnt3a for 4 h. Flag-CRBN immunoprecipitated from extracts of these cells were incubated with a mixture of recombinant GST-CK1α, ubiquitin, and a mixture of E1 and E2 ligases in the presence of vehicle or Mg-ATP, and incubated at 30 °C for 3 h. These reaction mixtures were subsequently analyzed by immunoblotting for the indicated proteins. A representative immunoblot (n = 3 independent experiments) is shown.