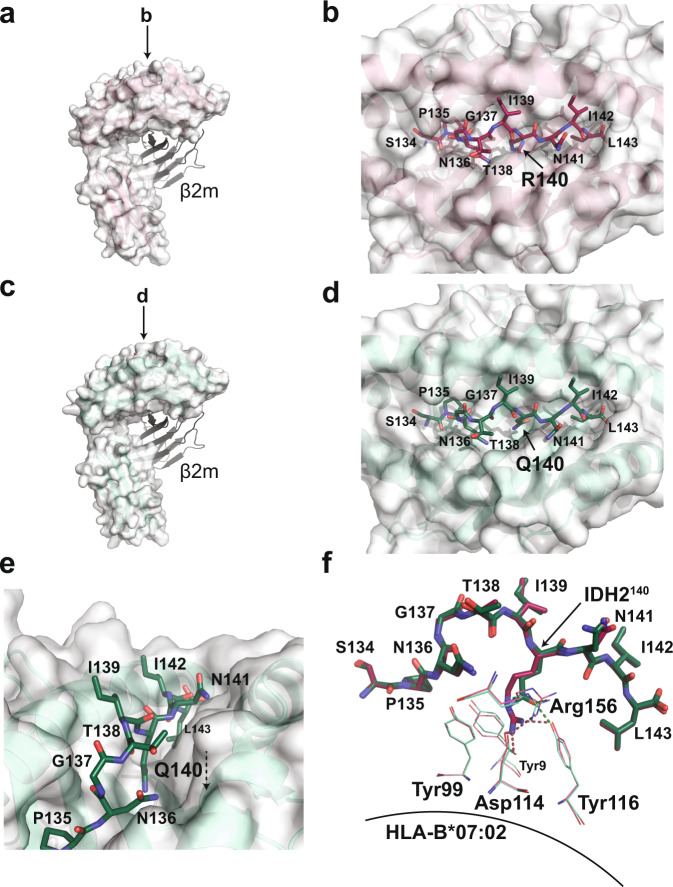
Fig. 2. The IDH2R140Q epitope is buried in the peptide-HLA-B*07:02 complex.
a Overall structure of the IDH2WT peptide bound to HLA-B*07:02 (PDB ID 6UJ8). The IDH2WT-HLA-B*07:02 complex is formed as a heterodimer between the heavy chain, represented as a white surface, and β2-microglobulin, represented as cartoon. The resolution of the IDH2WT-HLA-B*07:02 structure is 2.25 Å with an average B-factor of 40 Å2. b A bird’s-eye view of the IDH2WT peptide sitting in the HLA-B*07:02 peptide-binding groove. Amino acid residues of the IDH2WT peptide are labeled. c Overall structure of the IDH2R140Q peptide bound to HLA-B*07:02 (PDB ID 6UJ7). The heavy chain is represented as a white surface, and β2-microglobulin, represented as cartoon. The resolution of the IDH2R140Q-HLA-B*07:02 structure is 1.9 Å with an average B-factor of 34 Å2. d A bird’s-eye view of the IDH2R140Q peptide bound in the HLA-B*07:02 binding pocket. The residue of interest, IDH2R140Q, is buried in the peptide-binding groove. e Different orientation of (d) highlighting the downward position of the epitope of interest. f Structural alignment of the WT and mutant IDH2 peptides, showing the interactions between the epitope residue of interest and residues on the β-sheet floor of HLA-B*07:02. IDH2 peptide residues are labeled with single-letter amino acid codes. The HLA-B*07:02 residues are shown in green (mutant) and raspberry (WT) lines and labeled with three-letter amino acid codes; hydrogen bonds are represented as dotted lines.