FIGURE 3.
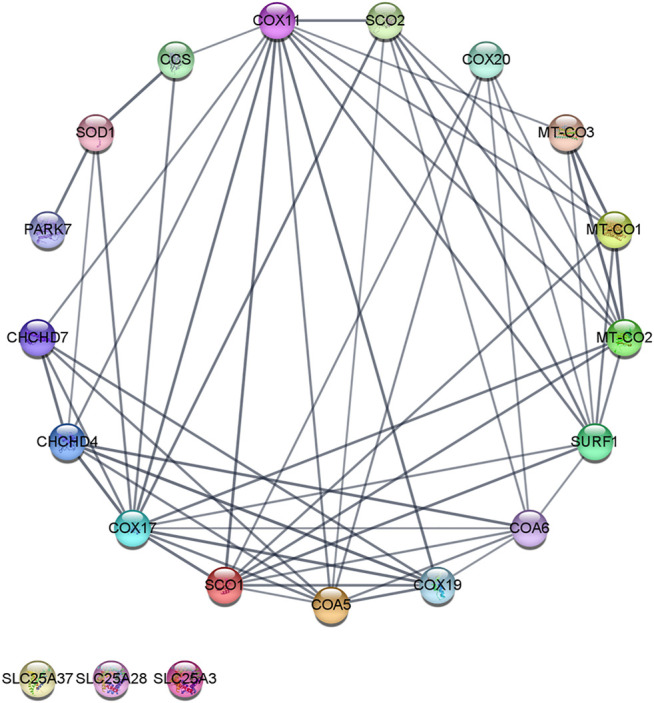
Copper protein network in mitochondria–STRING interaction network. The interaction network was created with the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database version 11.0. A medium confidence cutoff of 0, 7 was implemented in this work. The resulting protein association network for copper was visualized by a Cytoscape v3.8.2. Proteins are presented as nodes (circles) connected by lines (Edge) whose thickness represents the strength of the connection based on the STRING database. For example, in the network’s inferior part, three nodes (“SLC25A28,” “SLC25A37,” and “SLC25A37”) are not connected to the network. The node “CCS” is connected to “COX11,” “COX17,” and “SOD1” nodes. The edge (line) that connected “CCS” with “SOD1” is thicker than the edge between “CCS” and “COX11,” and nodes (shared named, Stringdb canonical name, display name, neighborhood connectivity—number of neighbors). COX17 (9606. ENSP00000261070, Q14061, COX17, 7); COX11 (9606. ENSP00000299335, Q9Y6N1, COX11, 7); SURF1 (9606. ENSP00000361042, Q15526, SURF1, 8); PET191 (9606. ENSP00000330730, Q86WW8, COA5, 8); SCO1 (9606. ENSP00000255390, O75880, SCO1, 8); MT-CO2 (9606. ENSP00000354876, P00403, MT-CO2, 8); COA6 (9606. ENSP00000355572, Q5JTJ3, COA6, 8); MIA40 (9606. ENSP00000295767, Q8N4Q1, CHCHD4, 8); COX19 (9606. ENSP00000342015, Q49B96, COX19, 9); SCO2 (9606. ENSP00000444433, O43819, SCO2, 9); MT-CO1 (9606. ENSP00000354499, P00395, MT-CO1, 9); COX23 (9606. ENSP00000306425, Q9BUK0, CHCHD7, 8); COX20 (9606. ENSP00000406327, Q5RI15, COX20, 9); SOD1 (9606. ENSP00000270142, P00441, SOD1, 9); MT-CO3 (9606. ENSP00000354982, P00414, MT-CO3, 9); CCS (9606. ENSP00000436318, O14618, CCS, 6); PARK7 (9606. ENSP00000418770, Q99497, PARK7, 4); SLC25A3 (9606. ENSP00000228318, Q00325, SLC25A3, 0); SLC25A28 (9606. ENSP00000359526, Q96A46, SLC25A28, 0); and SLC25A37 (9606. ENSP00000429200, Q9NYZ2, SLC25A37, 0). In Supplementary Material additional data of the copper protein interaction network can be found.
