Figure 2.
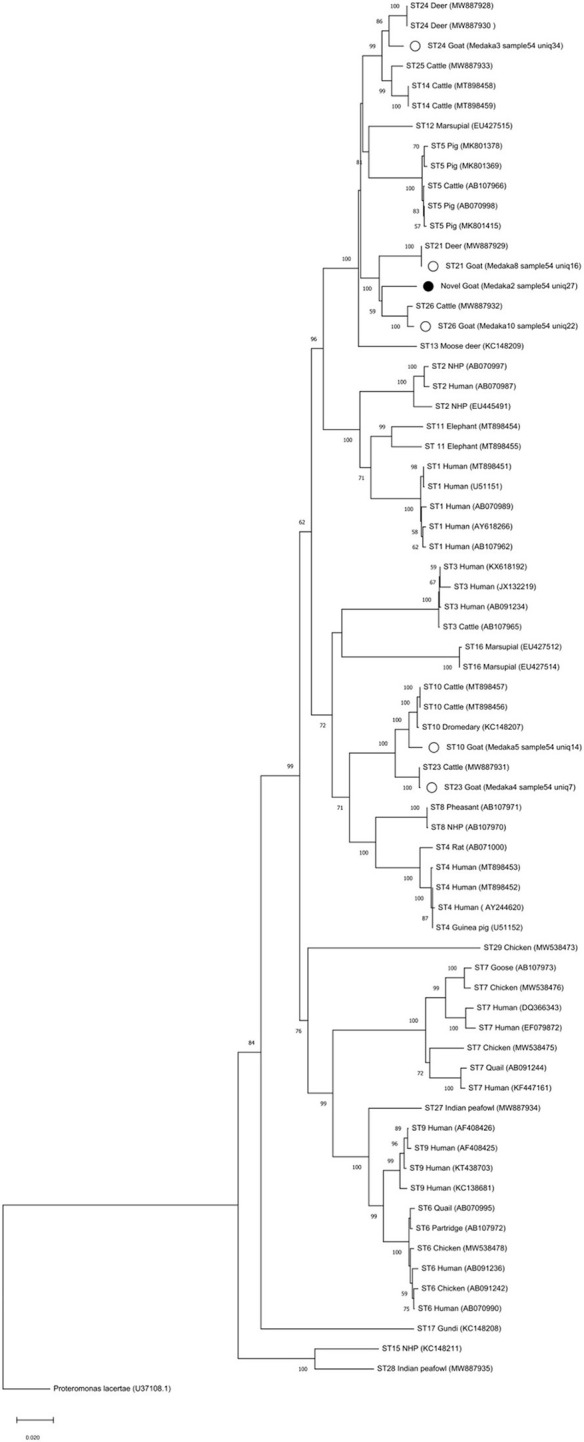
Phylogenetic reconstruction with MinION sequences. The full-length SSU rRNA gene nucleotide sequences obtained in this study were aligned with Blastocystis reference nucleotide sequences (accession numbers in parenthesis). Nucleotide sequences were aligned with the Clustal W algorithm and phylogenetic analysis was performed using the Neighbor-Joining (NJ) method and genetic distances calculated with the Kimura 2-parameter model using MEGA X (48, 49). Bootstrapping with 1,000 replicates was used to support the clades. The bootstrap number is shown on each node. Proteromonas lacertae was used as an outgroup. White circles show the samples with full-length obtained in this study and the black circle is showing the novel subtype found, called ST32.
