Figure 1. Phenotypic and genetic characterization of patient 1 with rare homozygous IFIH1 LoF variants.
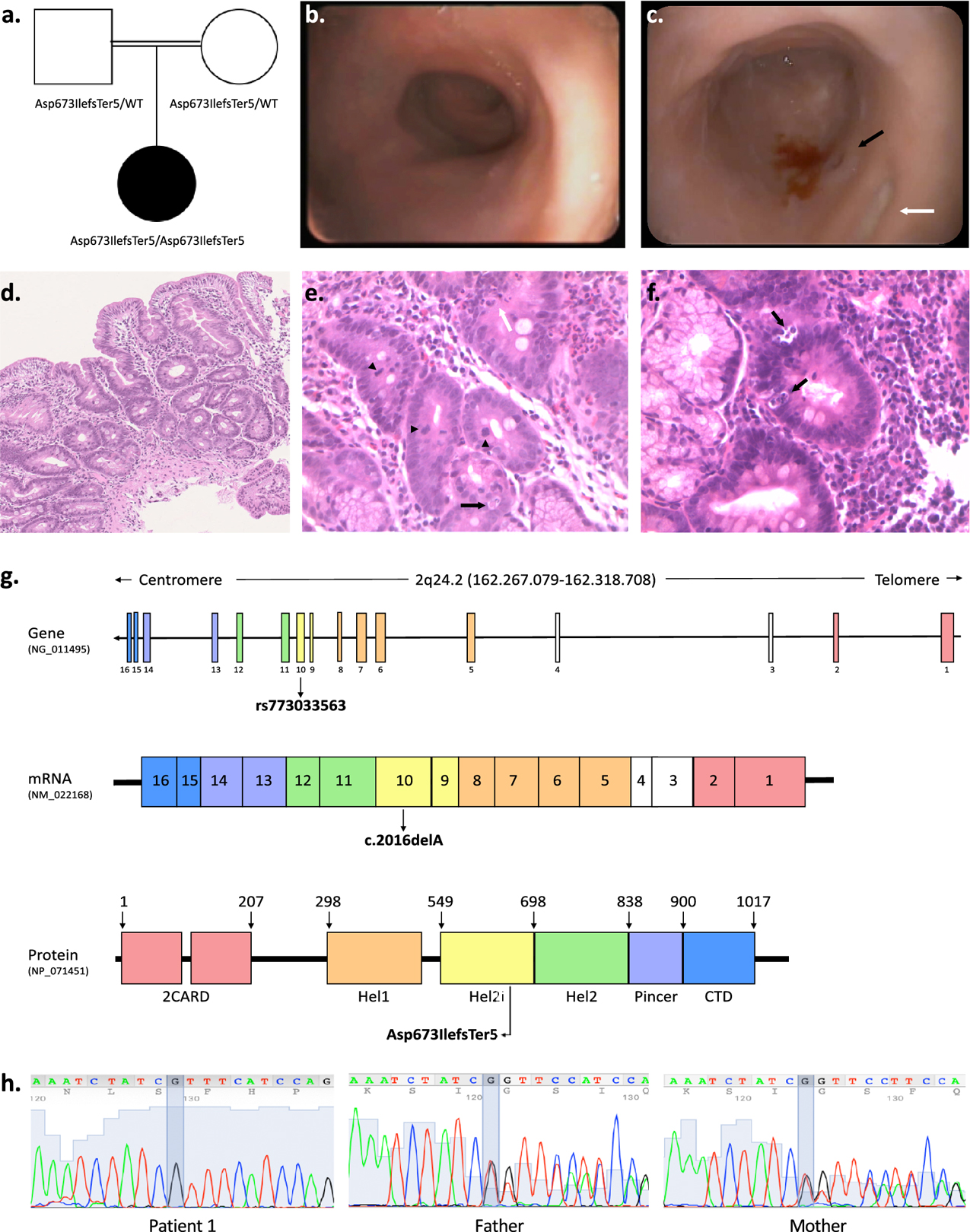
a. Pedigree of patient 1 indicating parental consanguinity and IFIH1 genotypes within the family. b. Upper gastrointestinal endoscopy showing absent duodenal folds with mucosal atrophy. c. Representative colonoscopy image of the left colon showing reduced haustral folds, a bleeding ulcer surrounded by mucosal inflammation (black arrow) and a fibrin-coated ulcer (white arrow). d-f. Duodenal mucosal histology (H&E; d 10x, e-f 20x) showing villous atrophy, crypt distortion, focal cryptitis (white arrow), mucin depletion, apoptotic bodies (black arrows), increased mitotic activity (arrow heads). g. Schematic of the human IFIH1 gene (NG_011495), mRNA (NM_022168) and protein (MDA5; NP_071451) showing the location of the homozygous frameshift deletion identified in patient 1 (rs773033563, c.2016delA, p. Asp673IlefsTer5). MDA5 is a 1025-residue protein (domain boundaries are marked with amino acid coordinates). The N-terminal of the protein contains two tandem caspase activation recruitment domains (2CARD) that activate MAVS. The central helicase domain is responsible for RNA-dependent ATP hydrolysis; it is made of two RecA-like domains (Hel1 and Hel2) and of the intervening Hel2i domain which facilitates the recognition of dsRNA. The Pincer domain is required for structural support and activation of the ATPase core of the protein. The C-terminal domain (CTD) is involved in dsRNA binding. h. Confirmatory Sanger sequencing demonstrating variant segregation within the family.
