Figure 2. Binding of CARD8-CT at Site B Which is Not Disrupted by DPP9 Inhibitors.
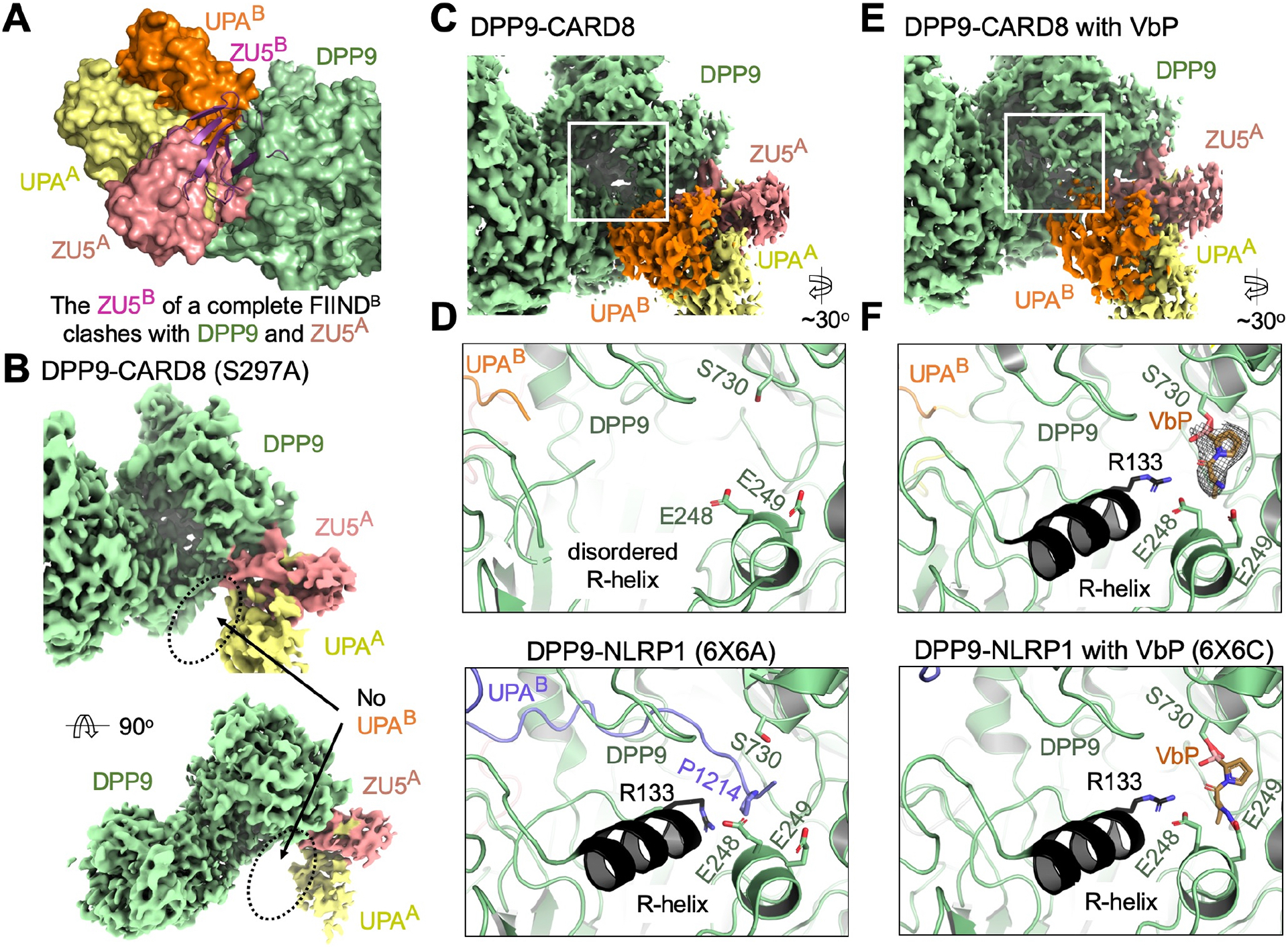
(A) Superimposition of the ZU5A-UPAA relationship onto UPAB to generate a theoretical ZU5B (purple). A theoretical ZU5B cannot be accommodated by the structure, explaining why site B is a dissociated UPAB, rather than a complete FIIND.
(B) Cryo-EM density of the DPP9-CARD8 (S297A) complex with no visible UPAB density. Autoproteolysis, and presumably, N-terminal degradation is required for site B association.
(C) Density of the DPP9-CARD8 complex near the DPP9 active site.
(D) Zoom-in of the DPP9 substrate tunnel in the DPP9-CARD8 complex. CARD8-UPAB binds near, but not into, the DPP9 substrate tunnel and the R-helix remains disordered. In comparison, zoom-in view of the NLRP1-DPP9 complex (PDB ID: 6X6A) at the DPP9 active site shows occupancy by the NLRP1-CT peptide.
(E) Density of the DPP9-CARD8 complex with bound VbP near the active site.
(F) Zoom-in of the DPP9 substrate tunnel in the DPP9-CARD8 complex with VbP. VbP binds near the active site, causing R-helix ordering, but does not displace CARD8-UPAB. in comparison, zoom-in view of the NLRP1-DPP9-VbP complex (PDB ID: 6X6C) at the DPP9 active site shows that VbP displaces the NLRP1-CT peptide.
