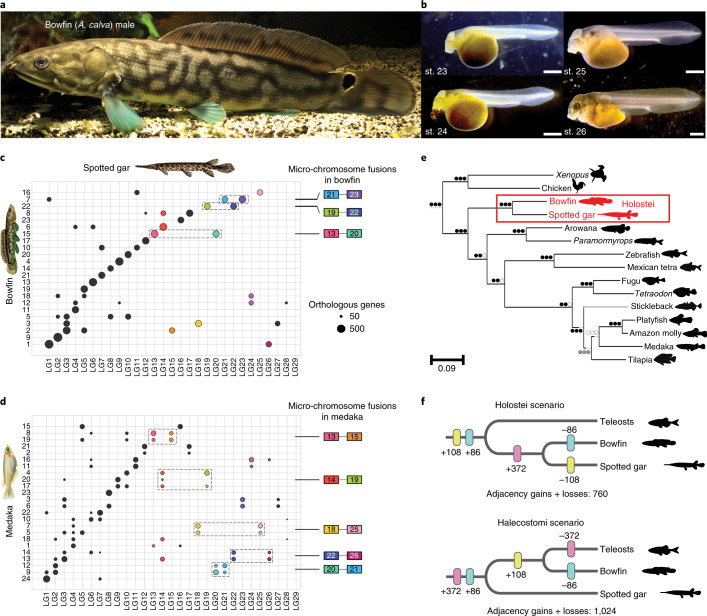
Fig. 1. Bowfin and the evolution of neopterygian genome organization.
a, Adult male bowfin. b, Bowfin stages (st.) 23–26 (ref. 53), covering critical phases of pectoral fin and gas bladder development (representative of n = 20 individuals per stage). Scale bar, 1 mm. c, Orthologies between bowfin and gar chromosomes and inferred bowfin micro-chromosome fusions for bowfin. d, Orthologies between medaka and gar chromosomes and inferred medaka micro-chromosome fusions. Circles, number of orthologous genes shared by bowfin and gar and/or medaka chromosomes (if in excess compared to random expectations). Ancestral micro-chromosomes are highlighted with colors; micro-chromosome fusions are indicated by dotted boxes. Colored boxes refer to the gar chromosome number. Micro-chromosome fusions differ between c and d: for example, medaka 9 and 12 result from a pre-TGD fusion of ancestral chromosomes orthologous to gar micro-chromosomes LG20 and LG21 and macro-chromosome LG2, followed by TGD duplication of the fusion chromosome (d). Bowfin 15 is a fusion of ancestral micro-chromosomes orthologous to gar LG13 and LG20; bowfin 7 is a fusion of ancestral chromosomes orthologous to gar LG1 and micro-chromosomes LG21 and LG23 (c). e, NJ phylogeny based on gene-order divergence built using a normalized breakpoint distance. Circles, bootstrap support for NJ, FastME and UPGMA analyses; black, 100%; gray ≥70%; white <70%. The Holostei clade is strongly supported (red box). Note that the location of the stickleback branch (gray) is in disagreement with the consensus phylogeny4,8 (but see ref. 85). f, Dollo parsimony applied to gains or losses of local gene adjacencies in Holostei (top) and Halecostomi (bottom) scenarios. Adjacencies shared by bowfin and gar only are in pink, those shared by bowfin and teleosts only are in yellow, and those shared by gar and teleosts only are in blue.