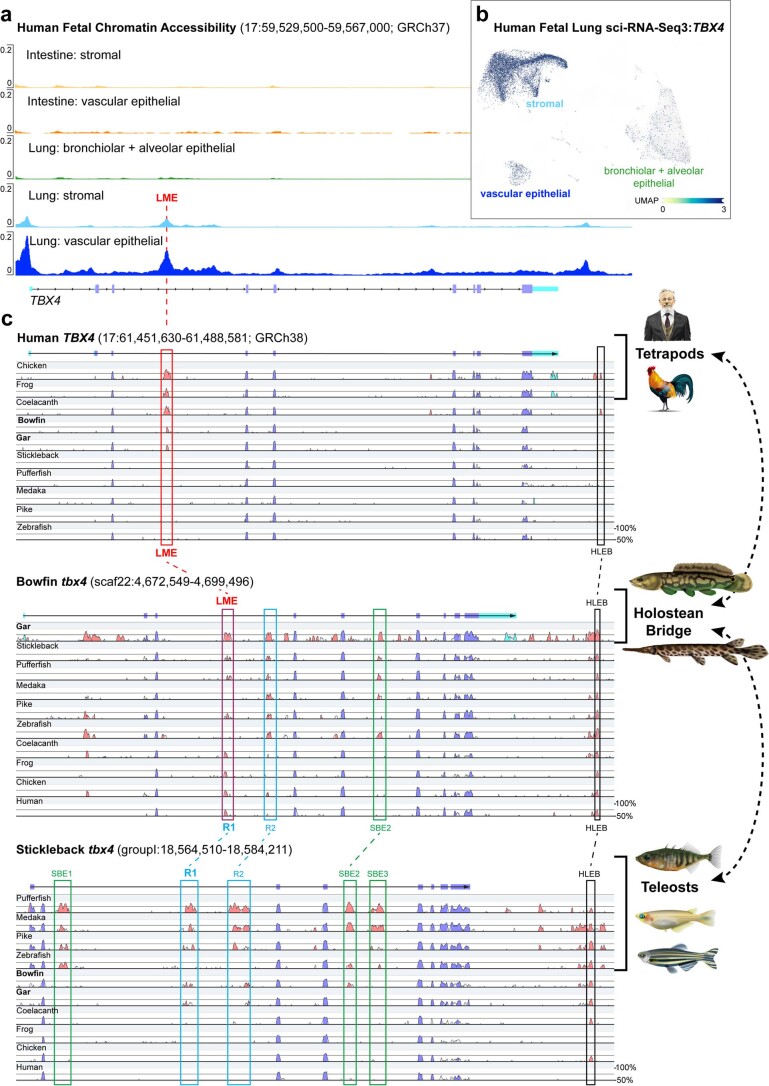
Extended Data Fig. 4. The holostean bridge connects vertebrate Tbx4 gene enhancers.
(a) Human fetal TBX4 chromatin accessibility shows a peak at the ‘lung mesenchyme enhancer’ (LME) in lung cells. (b) Human TBX4 single cell expression in fetal lung. See Supplementary Note 9 for details. (c) VISTA SLAGAN alignments with human (top), bowfin (middle), and stickleback (bottom) as reference. Top: Human TBX4 intron 3 ‘lung mesenchyme enhancer’ (LME, red box), driving expression in the developing mammalian lung bud60,61, shows conservation with other tetrapods, coelacanth, bowfin, and gar (and also with bichir12,126 and some sharks127, not shown), but no conservation with teleosts. HLEB: TBX4 hind limb enhancer B60 (black box). Bottom: The stickleback tbx4 gene contains three putative ‘swimbladder enhancers’86 (SBE1-3, green boxes). SBE2 is conserved with bowfin but not lobe-finned vertebrates. Intron 3 regions R1 and R2 (blue boxes) appear as conserved among neopterygians (teleosts, bowfin, gar), but not with coelacanth or tetrapods. HLEB (black box) is also present. Middle: The bowfin tbx4 gene identifies different conserved regions defined in the human- and stickleback-centric alignments (top, bottom): SBE2 (green box), R2, and HLEB. Lobe-finned LME and neopterygian R1 elements overlap within the bowfin genome, and are thus orthologous conserved non-coding elements, connected via the holosteans bridge (right). This bowfin LME/R1 region is also a bowfin developmental ncOCR (main text Fig. 4f, where it is shown on the reverse strand).