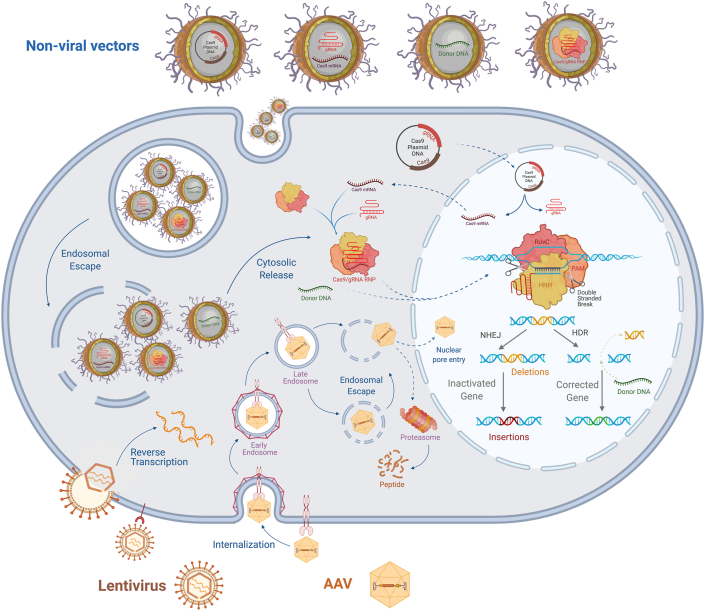
Figure 2.
CRISPR-Cas9 genome editing through viral or non-viral delivery. Representative depiction of mechanisms and strategies involved in CRISPR-Cas9 delivery with both viral and non-viral vectors. AAV and lentivirus both bind to cell surface receptors prior to cellular infection. Following cellular internalization, AAVs have the capacity to escape the endosomes and transport across the nuclear membrane prior to uncoating, though the capsid degradation mediated by proteasome can also occur in the cytoplasm. Following lentiviral cell membrane fusion is uncoating and release of its RNA contents, which then undergo reverse transcription to form complementary DNA. Non-viral vectors offer the advantage of carrying various forms of CRISPR-Cas9 cargoes including plasmid DNA, RNA, donor DNA, and RNP. Cellular entry of non-viral vectors is accomplished via endocytosis which requires the NP to escape these endosomes in order to carry out its intended genome editing. Following endosomal escape and cytosolic release, the cargo carried by a non-viral NP must travel to varying sites, such as the nucleus for transcription and/or cytoplasm for translation. Once necessary transcription and translation steps have taken place with nucleic acid delivery approaches, a RNP is formed and can translocate across the nuclear membrane for targeted genome editing. RNPs work to perform targeted DSBs by PAM- and sgRNA-mediated recognition of a specific sequence of chromosomal DNA. Once this recognition occurs, the Cas9 nuclease can perform a DSB utilizing its two nuclease domains the HNH and RuvC which cleave complementary and non-complementary DNA strands, respectively. Following a DSB, there are multiple fates for genome editing such as, but not limited to, NHEJ and HDR. NHEJ is utilized for genomic disruption or deletion, while HDR is utilized for gene correction, but requires the administration of an exogenous donor DNA template. AAV: adeno-associated virus, NP: nanoparticle, CRISPR: clustered regularly interspaced short palindromic repeats, Cas9: CRISPR-associated protein 9, RNP: ribonucleoprotein complexes, PAM: protospacer adjacent motif, DSB: double-stranded break, NHEJ: non-homologous end joining, HDR: homology-directed repair.