Figure 5 .
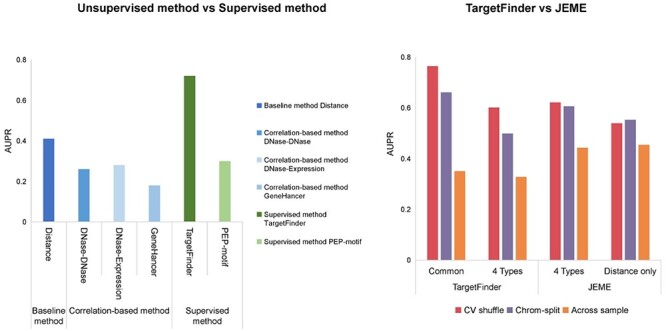
Performance of unsupervised and supervised methods. (A) Comparison of performance between distance-based, correlation-based and supervised methods using BENGI datasets. The AUPR scores of these six methods were evaluated by Moore et al [48]. (B) Comparison of regression-based model and trained classifier methods. Performances of TargetFinder and JEME on JEMEs ‘random target’ datasets using cross-validation with shuffling and chromosome-split strategies [82]. TargetFinder was validated with all features common in K562 and GM12878, and four epigenomic features (DNase-seq data and ChIP-seq data of H3K4me1, H3K27ac and H3K27me3) applied in JEME. JEME was validated with its four input features and distance feature, respectively. Across sample validation: TargetFinder and JEME were trained with K562 data and tested with GM12878 data. The AUPR scores of TargetFinder and JEME were evaluated by Cao and Fullwood [82].
