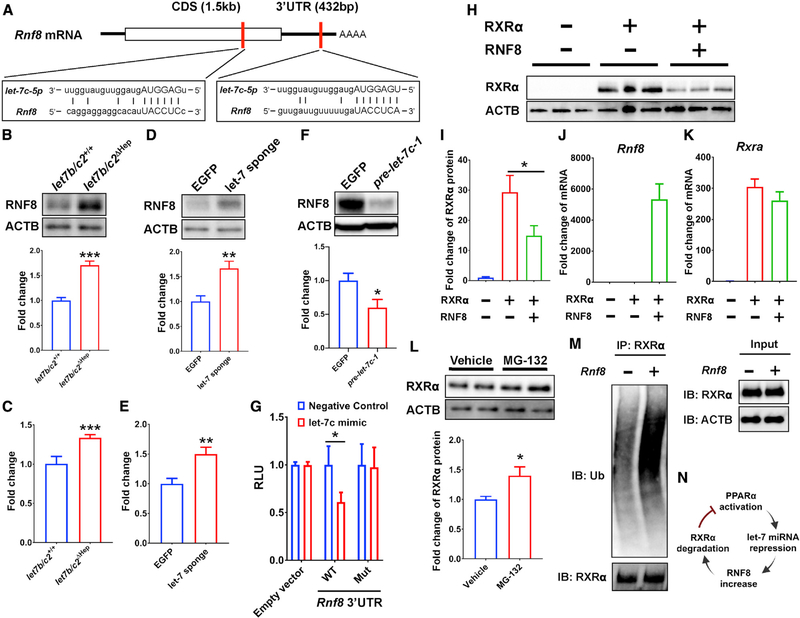
Figure 4. RNF8 is decayed by let-7 miRNA, and RXRα protein is ubiquitinated by RNF8 E3 ubiquitin ligase.
(A) Predicted let-7 miRNA binding sites in Rnf8 mRNA.
(B–F) Western blot analysis and densitometric quantification of RNF8 protein (B, D, and F) and qRT-PCR of Rnf8 mRNA (C and E) in let7b/c2+/+ and let7b/c2ΔHep (B and C); EGFP and let-7 sponge AAV-transduced (D and E); EGFP and pre-let-7c-1 AAV-transduced (F) livers treated with HFD feeding.
(G) 3′ UTR reporter assays in HepG2 cells transfected with Rnf8 wild-type or mutant 3′ UTR reporter constructs and a let-7c mimic expression vector.
(H and I) Western blot analysis (H) and densitometric quantification (I) of RXRα expression in Rxra- and Rnf8-transfected Hepa-1 cells.
(J and K) Fold change of Rnf8 (J) and Rxra (K) mRNA by qRT-PCR analysis in Rxra- and Rnf8-transfected Hepa-1 cells.
(L) Western blot analysis and the densitometric quantification of RXRα in Rxra- and Rnf8-transfected Hepa-1 cells treated with the proteasome inhibitor MG-132.
(M) Ubiquitination assays for Rxra- and Rnf8-transfected and MG-132-treated Hepa-1 cells. RXRα was immunoprecipitated and polyubiquitin detected by anti-ubiquitin antibody. RXRα expression was confirmed in whole-cell lysate as input.
(N) Scheme of 3-step inhibition for PPARα/RXRα pathway that the current study demonstrates.