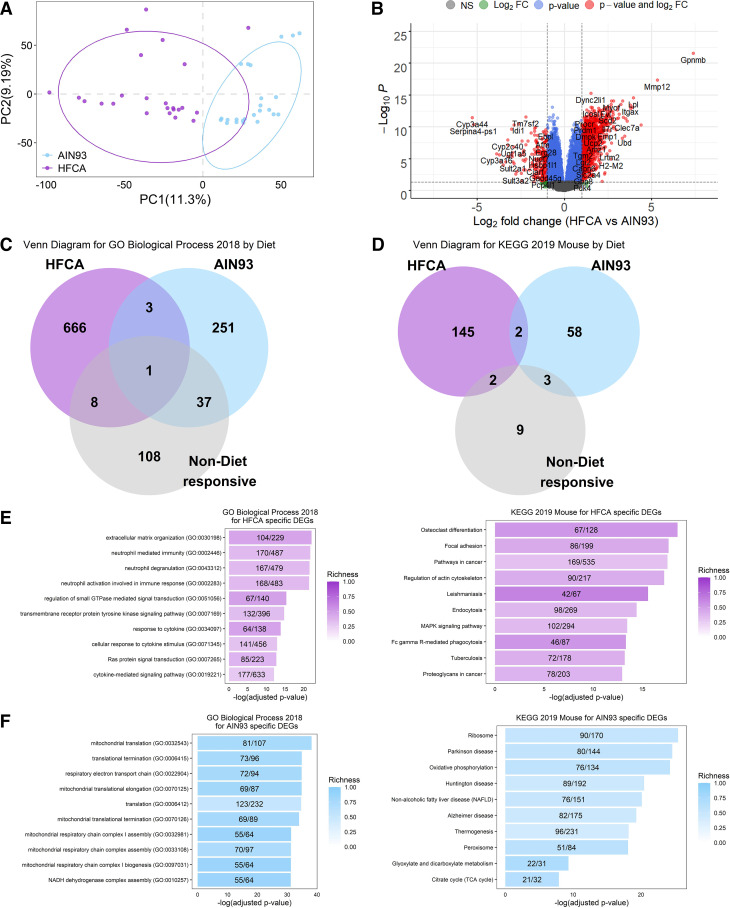
Figure 1.
Effect of diet on liver gene expression in female eight Collaborative Cross (CC) founder strains mice. We identified the effect of an atherogenic diet on global liver gene expression in eight CC founder strains (n = 48). Principal component (PC) analysis (A) and Volcano plot (B) between high-fat and cholic acid (HFCA) diet and AIN-93M diet in liver gene expression in eight CC founder strains. B: horizontal dotted lines indicate adj. P < 0.05, vertical dotted gray lines indicate a twofold difference. Venn diagram to identify overlapping gene ontology (GO) Biological Process 2018 terms (C) and Kyoto Encyclopedia of Genes and Genomes (KEGG) 2019 Mouse pathways (D) between upregulated genes in HFCA diet, upregulated genes in AIN-93M diet, or nondiet responsive genes in enrichment analysis. Top 10 GO terms and KEGG pathways of upregulated genes in HFCA diet (E) and AIN-93M diet (F) identified in enrichment analysis. Pathways were ordered from top to bottom by significance (highest to lowest) and colored by gene richness.