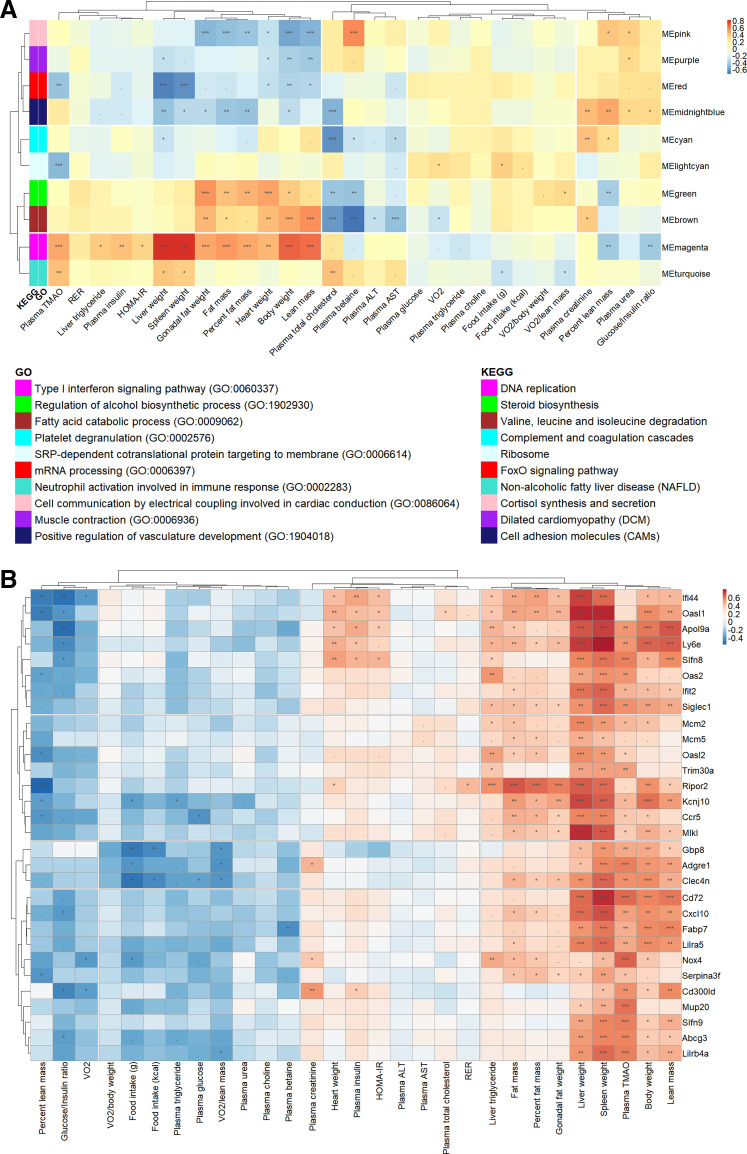
Figure 5.
Association of hepatic co-expression gene modules with metabolic traits in the eight Collaborative Cross (CC) founder strains in female mice. A: Spearman correlation between liver gene modules and metabolic traits in all mice. Module names were shown along the right axis, and top-enriched gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms in the legend. B: Spearman correlation between the top 30 high-fat and cholic acid (HFCA)-specific differential expression genes (DEGs) identified in the magenta module and metabolic traits in all mice. The P values were adjusted using the Benjamini–Hochberg (BH) false discovery rate (FDR) procedure. ***P < 0.001, **P < 0.01, *P < 0.05, .P < 0.10.