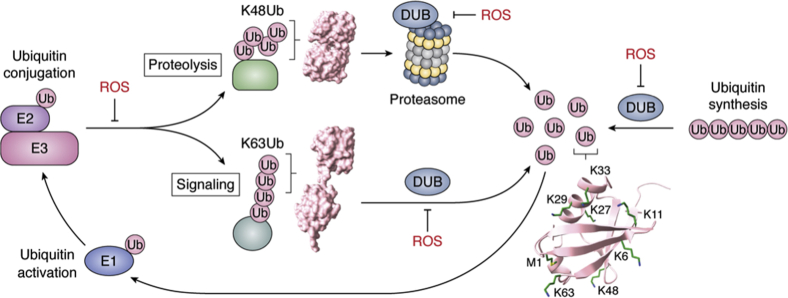
Figure 1.
Cycle of ubiquitin signaling. Ubiquitin (ub) is synthesized as polymers or fusions of ribosomal proteins that are cleaved into ubiquitin monomers by deubiquitinating enzymes (DUBs). Ubiquitin monomers are then used by E1 ubiquitin-activating enzymes to charge E2 ubiquitin conjugases, which work with or without E3 ubiquitin ligases to attach ubiquitin to targets. The conformation of K-linked ubiquitin chains (e.g., K48 or K63) determines the fate of the targets, whether they undergo ubiquitin signaling, or if they are sent to the proteasome for degradation. In either case, DUBs are responsible for removal of the ubiquitin and replenishment of the ubiquitin monomer pool. Reactive oxygen species (ROS)-sensitive steps are labeled in red. Structures depicted include ubiquitin (PDB: 1UBQ), and representatives of two forms of di-ubiquitin, K48 (PDB: 3AUL) and K63 (PDB: 3H7P).