Abstract
The evolutionarily ancient α/β hydrolase DWARF14-LIKE (D14L) is indispensable for the perception of beneficial arbuscular mycorrhizal (AM) fungi in the rhizosphere, and for a range of developmental processes. Variants of D14L recognise natural strigolactones and the smoke constituent karrikin, both classified as butenolides, and additional unknown ligand(s), critical for symbiosis and development. Recent advances in the understanding of downstream effects of D14L signalling include biochemical evidence for the degradation of the repressor SMAX1. Indeed, genetic removal of rice SMAX1 leads to the de-repression of symbiosis programmes and to the simultaneous increase in strigolactone production. As strigolactones are key to attraction of the fungus in the rhizosphere, the D14L signalling pathway appears to coordinate fungal stimulation and root symbiotic competency. Here, we discuss the possible integrative roles of D14L signalling in conditioning plants for AM symbiosis.
Keywords: Arbuscular mycorrhizal symbiosis, Alpha-beta hydrolase receptors, Strigolactone and karrikin, Gibberellic acid
Introduction
Arbuscular mycorrhizal (AM) symbiosis is a plant-fungal mutualism that arose approximately 450 million years ago in the early land plants [1,2]. The ability to engage in AM symbioses thereby predates the divergence of the plant lineages, and today involves the majority of the land plant species. The retention of the AM symbiosis to such extraordinary contemporary prevalence can likely be attributed to the significant nutritional benefit plants derive from the association, with up to 100% of phosphorus being delivered by the fungus as inorganic phosphate [3]. While it is long known that nitrogen is also transferred from the fungus to the plant [4], it was only recently shown that remarkably ~40% of overall plant nitrogen is symbiotically acquired in the form of nitrate [5∗∗]. This lends support for the AM symbiosis being the default plant nutrient uptake strategy, central to plant performance and ecosystem productivity, and the most important as well as most widespread symbiosis on earth (e.g. Refs. [6,7]).
The first step during the establishment of AM symbiosis is recognition between the plant and fungus in the rhizosphere. Plant roots release strigolactones (SLs), which not only draw the fungus to the root but also induce AM fungal metabolism, hyphal branching and production of fungal signalling compounds [8, 9, 10, 11, 12, 13, 14]. Conversely, fungal exudates include chitin-based signals [11,15], which are perceived by Lysin motif (LysM) receptor-like kinases (RLKs), and trigger the activation of the common symbiosis signalling pathway (CSSP) that in the legumes is equally required for endosymbiosis with nitrogen-fixing rhizobia. The fungus enters the outer cell layers of the root and proceeds to intracellularly colonise inner cortex cells, forming specialised structures called arbuscules. Arbuscules are the site where the bidirectional translocation of nutrients occurs. As an obligate biotroph, the fungus depends on plant-derived organic carbon, which amounts to ‘costs’ of ~20% of photosynthates for the plant [16]. Indeed, the plant host has the ability to control the establishment and extent of the interaction, as evidenced by well-fertilised plants, which display reduced levels of AM fungal colonisation (e.g. Refs. [17,18]).
In recent years, function of the rice α/β hydrolase (ABH) receptor DWARF14-LIKE (D14L) has been established as a strict requirement for enabling AM symbiosis, since mutations of D14L render rice insensitive to AM fungi [19]. The recent discovery that activation of the D14L signalling pathway leads to the de-repression of downstream genetic programmes by removal of a suppressor protein advances our mechanistic understanding of D14L function in symbiosis. Here, we propose that D14L signalling modulates the physiological condition of the plant to create a permissive state for AM symbiosis, underpinning attraction and enabling accommodation of the symbiotic fungus.
A rough guide to the D14L signalling pathway
Historically, D14L was first reported as a closely related homologue of D14 in the rice genome [20]. D14 back then was known to be required for SL sensitivity and later revealed as the rice SL receptor [20,21]. The Arabidopsis thaliana HYPOSENSITIVE TO LIGHT (HTL) described the first role for the D14L orthologue as being required for phytochrome and cryptochrome mediated de-etiolation in Arabidopsis [22]. Also, modulation of Arabidopsis seedling establishment upon exposure to the smoke constituent karrikin (KAR) depended on the same D14L homologue, prompting the additional name KARRIKIN INSENSITIVE 2 (KAI2) [23]. Arabidopsis KAI2 and rice D14L were shortly afterwards biochemically confirmed as KAR receptors [21,24].
The htl seedling photomorphogenesis phenotype [22] pointed early on towards the existence of an endogenous ligand (or multiple), which have tentatively been called KAI2 ligand (KL) [25,26], the significance of which was further supported by the discovery of diverse developmental roles for the KAI2 receptor in Arabidopsis, petunia and Lotus japonicus, including the regulation of cotyledon and rosette morphology [23], rosette shoot branching and leaf and petiole elongation [27], number of hypodermal passage cells [28∗∗], root hair density and length [29,30], root skewing [31], root system architecture [29, 30, 31], and also cuticle development [32].
KAR and SLs are structurally related butenolides that in the rhizosphere trigger seed germination of many fire-following and parasitic plants, respectively (for a recent review see Ref. [33]). Evidence for D14L receptors mediating the response to KAR in the fire-prone Brassica tournefortii was obtained by B. tournefortii homologues overcoming seed dormancy in Arabidopsis [34∗]. Interestingly, a fast-evolving clade of duplicated and diversified (‘d’ clade) D14L receptors from the parasitic plant Striga hermonthica were responsible for the critical perception of host-released SL to induce seed germination, whereas treatment with KAR had no germination-promoting effect [35]. In contrast, cross-species complementation revealed that conserved (‘c’ clade) HTL paralogues from different parasitic plant species restored Arabidopsis kai2 phenotypes, indicative of their ability to recognise KL [25]. KL therefore carries the hallmarks of a plant hormone with its receptor regulating responses to a variety of endogenous developmental but also environmental cues.
Through genetic approaches in Arabidopsis it was shown that activation of the KAI2 receptor leads to the recruitment of an F-box protein of the Skp1-Cullin-F-box (SCF) complex, MORE AXILLARY GROWTH2 (MAX2), and the removal of the negative regulators SUPPRESSOR OF MAX2 1 (SMAX1) and SMAX1-LIKE2 (SMXL2) that operate downstream of KAI2 and MAX2 and control partly overlapping developmental processes [36, 37, 38]. Intriguingly, the SL receptor D14 signals through the same F-box protein, inducing the polyubiquitination and degradation of distinct SMXL proteins [36,38, 39, 40]. Such extraordinary relatedness likely results from the shared evolutionary history with D14L being the older paralogue to D14 [41,42]. The long-awaited biochemical evidence for MAX2-dependent polyubiquitination and proteolysis also of SMAX1/SMXL2 upon receptor activation was recently obtained [43∗,44∗]. Interestingly, removal of SMXL2 could be triggered by interaction with either D14 or KAI2 depending on the nature of the synthetic ligand, whereas SMAX1 degradation occurred specifically upon activation of KAI2 signalling [44∗]. However, degradation of SMAX1/SMXL2 was still occurring in max2 mutants, arguing for an elusive MAX2-independent mechanism, and revealing an unanticipated high complexity of signalling fine-tuning [43∗]. In addition, direct interaction between KAI2 and SMAX1/SMXL2 was demonstrated, offering an explanation for the previously reported degradation of the KAI2 receptor upon ligand treatment, which was also MAX2-independent, and suggested turnover of the receptor after having signalled, the purpose of which remains obscure [43∗,44∗,45].
An equivalent signalling pathway operates in the control of rice mesocotyl length [19,46], with D14L shown to form a complex with the MAX2 orthologue DWARF3 (D3) and SMAX1, resulting in ubiquitination and proteolysis of SMAX1 [47∗∗]. SMAX1 also interacted with a member of the transcriptional corepressors TOPLESS-RELATED-PROTEIN2 (TPR2) involving the ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif. Although the functional relevance of TPR2 for mesocotyl elongation has not been assessed, the SMAX1-TPR2 interaction suggests that at least a proportion of the D14L controlled genes might be regulated through the release of TPR2 repression by removal of SMAX1 [47∗∗].
D14L signalling in AM symbiosis
In addition to being central to developmental processes, rice D14L is essential for the establishment of AM symbiosis, as the interaction is completely abolished in the rice d14l mutant [19]. Consistent with a role in perceiving AM fungi, the d14l mutant failed to mount a diagnostic transcriptional response to AM fungal signals [19]. Mutations of D3 also showed severe defects in AM colonisation, suggesting that other known components of the D14L pathway are required for AM symbiosis [19,48,49∗∗]. Characterisation of the interaction of rice smax1 with AM fungus Rhizophagus irregularis revealed elevated colonisation levels, thereby identifying SMAX1 as a negative regulator of AM symbiosis [49∗∗] (Table 1). Double mutants of d14l/smax1 and d3/smax1 confirmed that, in addition to development signalling, SMAX1 also operates downstream of the D14L receptor and the D3 F-box protein in symbiosis signalling. Transcriptome analysis of smax1 and d3/smax1 found several members of the CSSP to be upregulated in a smax1 mutant-dependent fashion. The significantly stronger mycorrhizal phenotype of d14l rice mutants relative to individual CSSP mutants might plausibly be explained by collective interference with the function of multiple CSSP components [19,49∗∗,50]. The epistatic relationship between the D14L and the CSSP is currently unknown and awaits further investigation.
Table 1.
List of D14L, SL, and GA-related genes whose involvement in AM symbiosis has been shown by genetic analysis.
| Gene | Plant species | AM colonisation | Function | Reference(s) |
|---|---|---|---|---|
| D14L | Rice | – | ABH required for D14L signalling | [19] |
| KAI2 | Petunia hybrida | – | [28] | |
| D3 | Rice | – | F-box protein required for D14L and SL signalling | [48] |
| RMS4 | Pisum sativum | ++ | [57] | |
| MAX2A | Petunia hybrida | – | [28] | |
| SMAX1 | Rice | ++++ | SMXL/D53 family suppressor of D14L signalling | [49] |
| D14 | Rice | ++++ | ABH required for SL signalling | [19,48] |
| D10 | Rice | ++ | SL biosynthesis | [12,58] |
| D17 | Rice | ++ | [12,48,58] | |
| RMS1/CCD8 | Pisum sativum | ++ | [17,53,57,59] | |
| CCD7 | Solanum lycopersicum | ++ | [60] | |
| CCD8 | Solanum lycopersicum | ++ | [61] | |
| GID1 | Rice | +++ | ABH required for GA signalling | [62] |
| SLR1 | Rice | ++ | GA signalling repressor(s) | [55,62] |
| DELLA1/2 | Medicago truncatula | + | [63] | |
| LA and CRY | Pisum sativum | ++ | [64] | |
| PRO | Solanum lycopersicum | ++ | [65] | |
| NA | Pisum sativum | ++++ | GA biosynthesis | [64] |
++++, high AM colonisation; +++, wild-type AM colonisation; ++, reduced AM colonisation but normal infection structures or weak defects; +, low AM colonisation with strong defects in infection structures; -, no AM colonisation. D14L, Dwarf14-like; KAI2, Karrikin-insensitive2; D3, Dwarf3; RMS4, Ramosus4; MAX2A, More Axillary Growth2A; SMAX1, Suppressor of Max2 1; D14, Dwarf14; D10, Dwarf10; D17, Dwarf17; RMS1, Ramosus1; CCD8, Carotenoid Cleavage Dioxygenase8; CCD7, Carotenoid Cleavage Dioxygenase7; GID1, Gibberellin Insensitive Dwarf1; SLR1, Slender Rice1; PRO, Procera; NA, Kaurenoic acid oxidase.
Importantly, the D14L signalling pathway appears to function as a master control mechanism that can switch plant competency for symbiosis into an ‘on’ or ‘off’ condition. This raises the interesting question whether the plant is by default in an ‘off’ state, requiring induction, or is usually rather in an ‘on’ state that can, however, be stopped when the symbiosis is not needed, for example in well-fertilised plants. Following this, the question about the nature and origin of the ligand then arises. Conceivably, either AM fungi might release a butenolide with structural similarity to KAR, or alternatively, an endogenous plant ligand might be produced whose detection creates a permissive state for symbiosis [19]. Although the first scenario cannot be excluded as fungi are capable of producing butenolides (e.g. Ref. [51]), we favour the second hypothesis, which is supported by the inability to detect SMAX1 in untreated wild-type rice material, thus indicative of activated D14L signalling [47∗∗,49∗∗]. Also, treating rice roots with KAR did not enhance mycorrhizal colonisation, and did not lead to significant changes in the root transcriptome [19], possibly reflecting that KAR is not perceived by rice roots. Alternatively however, already activated D14L signalling might fail to be further activated. The first hypothesis seems unlikely as Arabidopsis and L. japonicus root developmental or skewing properties are altered by KAR in a D14L dependent fashion [30,31,52∗]. Resolving these alternatives requires a more comprehensive analysis with a variety of natural and synthetic ligands.
The mechanisms that underpin specificity of D14L signalling in the control of mesocotyl length or symbiosis are unknown but could be brought about by different KLs present in the different type of tissues. Indeed, comparing hypocotyl with root tissue, L. japonicus responded differentially to different synthetic KLs in an organ-specific way [52∗]. Alternatively, SMAX1 targets possibly vary in the different tissue contexts causing specific transcriptional readouts. The transcriptional corepressor TPR2 might be a first candidate as it interacts with SMAX1 in rice mesocotyl development [47∗∗] but its functional relevance for symbiosis establishment requires experimental verification.
D14L signalling integrates SL and likely also GA biosynthesis
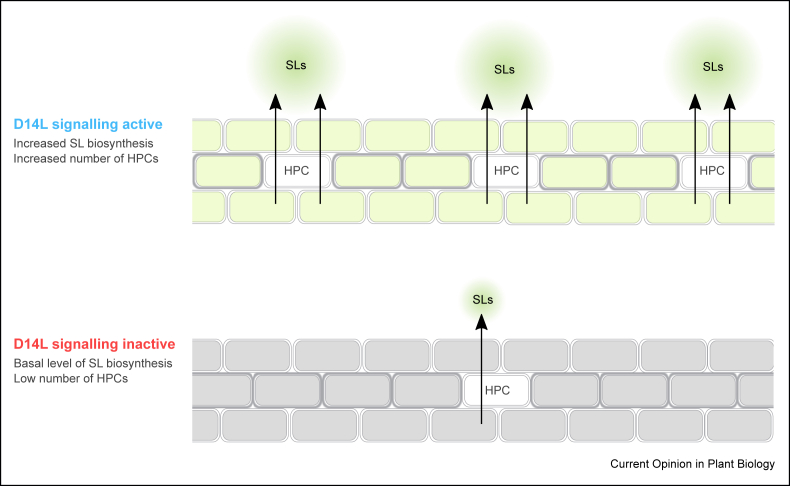
The smax1 and d3/smax1 transcriptome analysis also revealed that removal of SMAX1 causes induction of SL biosynthesis, which was mirrored by significantly elevated SL levels in rice roots [49∗∗]. This was more pronounced in d3/smax1 as compared to smax1. Consistently, d3/smax1 also showed a tendency for higher fungal colonisation than smax1, which was not seen in d14l/smax1 double mutant, the latter being specifically defective in D14L but not SL signalling. This lends a hypothesis that D3-mediated SL signalling provides a layer of regulation in AM symbiosis, synergistically to D14L signalling. As SLs are important rhizosphere signals for the attraction of the fungus, D14L signalling might integrate ‘calling for the fungus’ via SL production with conditioning the plant for AM symbiosis. In Petunia hybrida hypodermal passage cells (HPCs) promote the release of SL into the rhizosphere via the ABC transporter PDR1 [53]. Interestingly, the abundance of HPCs is negatively regulated by SL signalling but promoted by D14L signalling, as evidenced by the decreased number of HPCs in petunia kai2a mutants where SL signalling is active [28∗∗]. Conceivably, D14L might establish a link between increased SL production [45∗∗] and facilitated export of SL substrate through altering cell differentiation [28∗∗]. How far similar mechanisms are at play in rice is currently unknown but can perhaps be anticipated, as petunia kai2a mutants reproduced the strong AM symbiosis phenotype of rice d14l, reflected by the complete absence of AM fungal colonisation [28∗∗]. In summary, in a hypothetical model, petunia D14L signalling might simultaneously increase the level of SL and the availability of its ‘gates’ for enticing the fungus towards the root, while at the same time creating a permissive state for symbiosis (Figure 1). Related strategies might exist across a broader range of plant species.
Figure 1.
Hypothetical model for the proposed link between D14L signalling leading to increased SL production and number of hypodermal passage cells for SL export in petunia. When D14L signalling is active, SL biosynthesis and HPC density are increased [28∗∗,49∗∗], resulting in increased levels of SLs being exuded into rhizosphere. In the absence of D14L signalling, a lower number of HPCs and the basal level of SL biosynthesis results in lower levels of SLs being exported out of the root.
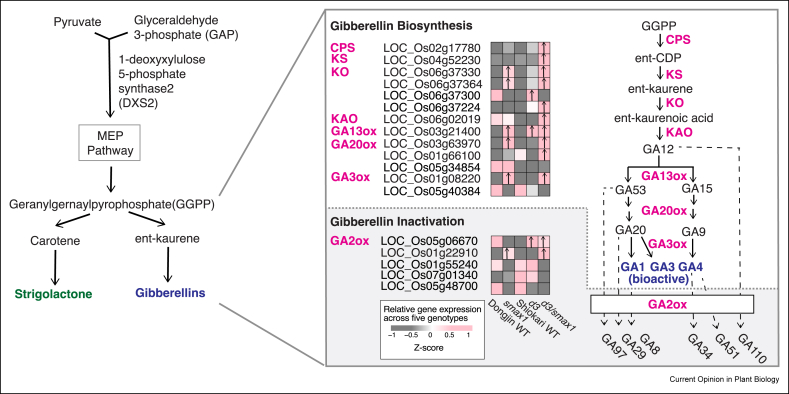
SL and gibberellic acid (GA) are both derived from geranylgeranyl pyrophosphate (GGPP) produced in the MEP pathway, after which the pathways diverge with SL being produced via carotene and GA via ent-kaurene (Figure 2). In addition to the induction of the SL biosynthesis pathway, the rice smax1 and d3/smax1 transcriptome data revealed a signature consistent with the upregulation of the GA biosynthetic pathway from the first committed step mediated by copalyl pyrophosphate synthase (CPS) to the final gibberellin 3-oxidase (GA3ox) that gives rise to bioactive GAs [49∗∗] (Figure 2). This signature was particularly pronounced in the double mutant d3/smax1, which additionally integrates the effects of defective SL signalling. Transcript levels of GA inactivating gibberellin 2-oxidase were also elevated in smax1 and d3/smax1 mutants, suggesting the fine-tuned adjustment of GA levels. Although GA levels were not quantified, these transcriptome data point towards the D14L signalling pathway regulating the biosynthesis of GA concomitant with SL. The effect of GA on AM symbiosis development is complex with reported positive and negative effects on fungal root colonisation at later stages of the interaction (for a recent review see Ref. [54]), while a requirement during presymbiosis has not been reported. In rice, GA is known to inhibit SL biosynthesis leading to reduced infection by the parasitic plant S. hermonthica [55]. The parallel induction of SL and GA biosynthetic pathways may indicate a feedback mechanism to achieve a favourable balance of SL levels. However, an SL-dependent overriding effect of the usually dominant role of GA signalling in seed germination was recently observed in Arabidopsis expressing S. hermonthica D14L (HTL) receptors, demonstrating that repressed GA-signalling can be bypassed by activating D14L signalling [56∗]. Clearly, the interplay between D14L and GA signalling comes in a variety of flavours that require further investigation, starting from the quantification of GA levels in D14L signalling mutants.
Figure 2.
Gene expression of GA metabolic pathway genes in D14L signalling mutants. GA and SL biosynthesis result from biosynthetic pathways that share initial steps but diverge after geranylgeranyl pyrophosphate (GGPP) (left). The metabolic pathway for gibberellic acid (right). Genes that were upregulated in either smax1, d3 or smax1/d3 were selected and the normalised expression values across six genotypes are colour coded according to the z-score [49∗∗]. Genes with expression levels higher in either smax1, d3 or d3/smax1 mutants relative to their respective wild types (WTs) are indicated with upward arrows inside the box (fold change <1.5, FDR <0.05). Dongjin WT and Shiokari WT refer to cultivars in which smax1 and d3 arose, respectively. Solid and dotted lines represent synthesis and inactivation steps, respectively. MEP, 2-methyl-erythritol-4-phosphate; CPS, copalyl pyrophosphate synthase; ent-CDP, ent-copalyl diphosphate; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase; GA, gibberellic acid/gibberellin; GA13ox, gibberellin 13-oxidase; GA20ox, gibberellin 20-oxidase; GA3ox, gibberellin 3-oxidase; GA2ox, gibberellin acid 2-oxidase.
Concluding remarks
Here, we discuss that D14L signalling is a central regulator of symbiotic competency, mechanistically linked with the biosynthesis of SL, the best known and potent attractant for AM fungi (Figure 3). We propose that D14L signalling in rice roots under normal conditions is activated by the availability of an endogenous ligand but can be suppressed under non-favourable circumstances through accumulation of the SMAX1 repressor, which directly or indirectly blocks the transcription of symbiosis programmes (Figure 3). At present, it is unknown whether AM symbiosis regulation is an ancestral or derived trait for D14L, since the symbiotic function of homologues from distantly related plants have not yet been examined. The role of D14L in AM symbiosis is however conserved in the angiosperms, as D14L is required for AM symbiosis in both the eudicot petunia [28∗∗] and the monocot rice [19,49∗∗]. Further research into crosstalk between presymbiotic phytohormones, the identity of a D14L ligand, the downstream signalling pathway of SMAX1, and the conservation of a symbiotic role of D14L signalling will collectively provide us with the information necessary to understand how D14L conditions plants for symbiosis.
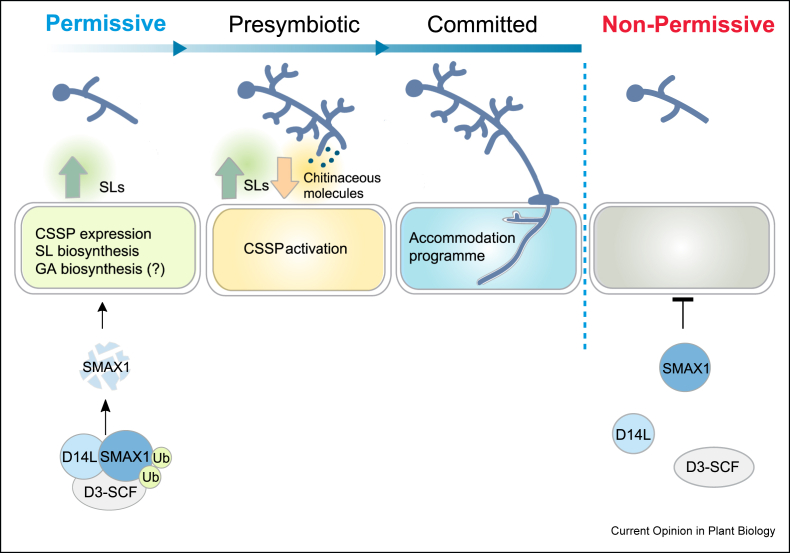
Figure 3.
Model for D14L signalling in conditioning of plants for AM symbiosis. D14L signalling leads to the removal of the repressor SMAX1 and to the upregulation of the expression of CSSP components, increased SL biosynthesis, and increased expression of GA biosynthesis genes. This D14L activity creates a permissive state of the plant for AM symbiosis, setting the scene for presymbiosis signalling with AM fungi. During presymbiosis, the biochemical communication between the plant and the AM fungus activates the CSSP in preparation for accommodation. Once committed, formation of penetrative structure, the hyphopodium, occurs and the activated accommodation programme enables the establishment of AM symbiosis. However, in the absence of D14L signalling, the accumulation of SMAX1 results in a non-permissive condition for AM symbiosis development.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
R.H. is funded by the Biotechnology and Biological Sciences Research Council (BBSRC) Doctoral Training Partnership at the University of Cambridge. J.C. is supported by Engineering the Nitrogen Symbiosis for Africa (ENSA), which is funded by the Bill & Melinda Gates Foundation (BMGF) as OPP1028264. The authors thank Edwin Jarratt Barnham and Gizem Kiyak for useful feedback on an earlier version of the manuscript.
This review comes from a themed issue on Biotic interactions
Edited by Jeffery L. Dangl and Jonathan D. G. Jones
References
- 1.Remy W., Taylor T.N., Hass H., Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. Dec. 1994;91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heckman D.S. Molecular evidence for the early colonization of land by fungi and plants. Science. Aug. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 3.Smith S.E., Smith F.A., Jakobsen I. Functional diversity in arbuscular mycorrhizal (AM) symbioses: the contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol. May. 2004;162:511–524. doi: 10.1111/j.1469-8137.2004.01039.x. [DOI] [Google Scholar]
- 4.Govindarajulu M. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature. Jun. 2005;435:819–823. doi: 10.1038/nature03610. [DOI] [PubMed] [Google Scholar]
- Wang S. Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc Natl Acad Sci USA. Jul. 2020;117:16649–16659. doi: 10.1073/pnas.2000926117. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work shows that a significant proportion, ~40%, of overall nitrogen uptake in rice is symbiotically derived and unexpectedly transferred to the plant in the form of nitrate.
- 6.Brachmann A., Parniske M. The most widespread symbiosis on earth. PLoS Biol. Jul. 2006;4:e239. doi: 10.1371/journal.pbio.0040239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutjahr C., Parniske M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol. Oct. 2013;29:593–617. doi: 10.1146/annurev-cellbio-101512-122413. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama K., Matsuzaki K., Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. Jun. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 9.Besserer A. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. Jun. 2006;4:e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besserer A., Bécard G., Jauneau A., Roux C., Séjalon-Delmas N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. Sep. 2008;148:402–413. doi: 10.1104/pp.108.121400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genre A. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca 2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. Apr. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 12.Kobae Y. Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol. 2018 doi: 10.1093/pcp/pcy001. [DOI] [PubMed] [Google Scholar]
- 13.Parniske M. Cue for the branching connection. Nature. Jun. 2005;435:750–751. doi: 10.1038/435750a. [DOI] [PubMed] [Google Scholar]
- 14.Tsuzuki S., Handa Y., Takeda N., Kawaguchi M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. MPMI. Apr. 2016;29:277–286. doi: 10.1094/MPMI-10-15-0234-R. [DOI] [PubMed] [Google Scholar]
- 15.Maillet F. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. Jan. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsen I., Rosendahl L. Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber plants. New Phytol. May. 1990;115:77–83. doi: 10.1111/j.1469-8137.1990.tb00924.x. [DOI] [Google Scholar]
- 17.Breuillin F. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning: phosphate and Petunia mycorrhiza development and functioning. Plant J. Dec. 2010;64:1002–1017. doi: 10.1111/j.1365-313X.2010.04385.x. [DOI] [PubMed] [Google Scholar]
- 18.Bonneau L., Huguet S., Wipf D., Pauly N., Truong H.-N. Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol. Jul. 2013;199:188–202. doi: 10.1111/nph.12234. [DOI] [PubMed] [Google Scholar]
- 19.Gutjahr C. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science. Dec. 2015;350:1521–1524. doi: 10.1126/science.aac9715. [DOI] [PubMed] [Google Scholar]
- 20.Arite T. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. Aug. 2009;50:1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- 21.Kagiyama M. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Gene Cell. Feb. 2013;18:147–160. doi: 10.1111/gtc.12025. [DOI] [PubMed] [Google Scholar]
- 22.Sun X.-D., Ni M. HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol Plant. Jan. 2011;4:116–126. doi: 10.1093/mp/ssq055. [DOI] [PubMed] [Google Scholar]
- 23.Waters M.T. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. Apr. 2012;139:1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- 24.Guo Y., Zheng Z., La Clair J.J., Chory J., Noel J.P. Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis. Proc Natl Acad Sci USA. May. 2013;110:8284–8289. doi: 10.1073/pnas.1306265110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conn C.E., Nelson D.C. Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci. Jan. 2016;6 doi: 10.3389/fpls.2015.01219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y.K., Flematti G.R., Smith S.M., Waters M.T. Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Front Plant Sci. Dec. 2016;7 doi: 10.3389/fpls.2016.01799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X. Arabidopsis SMAX1 overaccumulation suppresses rosette shoot branching and promotes leaf and petiole elongation. Biochem Biophys Res Commun. May 2021;553:44–50. doi: 10.1016/j.bbrc.2021.03.006. [DOI] [PubMed] [Google Scholar]
- Liu G. Strigolactones play an important role in shaping exodermal morphology via a KAI2-dependent pathway. iScience. Jul. 2019;17:144–154. doi: 10.1016/j.isci.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article reports that the number of hypodermal passage cells for SL export in petunia is negatively and positively regulated through D14 and D14L signalling, respectively.
- 29.Villaécija-Aguilar J.A. SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet. Aug. 2019;15 doi: 10.1371/journal.pgen.1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carbonnel S., Das D., Varshney K., Kolodziej M.C., Villaécija-Aguilar J.A., Gutjahr C. The karrikin signaling regulator SMAX1 controls Lotus japonicus root and root hair development by suppressing ethylene biosynthesis. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.2006111117. p. 202006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swarbreck S.M., Guerringue Y., Matthus E., Jamieson F.J.C., Davies J.M. Impairment in karrikin but not strigolactone sensing enhances root skewing in Arabidopsis thaliana. Plant J. 2019;98:607–621. doi: 10.1111/tpj.14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W. The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet. Nov. 2017;13 doi: 10.1371/journal.pgen.1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao J., Waters M.T. Perception of karrikins by plants: a continuing enigma. J Exp Bot. Mar. 2020;71:1774–1781. doi: 10.1093/jxb/erz548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.K. Divergent receptor proteins confer responses to different karrikins in two ephemeral weeds. Nat Commun. Dec. 2020;11:1264. doi: 10.1038/s41467-020-14991-w. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors confirmed that KAI2 genes from the fire follower Brassica tournefortii mediate KAR recognition leading to seed germination and identified residues conditioning ligand selectivity.
- 35.Conn C.E. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science. Jul. 2015;349:540–543. doi: 10.1126/science.aab1140. [DOI] [PubMed] [Google Scholar]
- 36.Nelson D.C. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. May 2011;108:8897–8902. doi: 10.1073/pnas.1100987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanga J.P., Smith S.M., Briggs W.R., Nelson D.C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. Sep. 2013;163:318–330. doi: 10.1104/pp.113.221259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soundappan I. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell. Nov. 2015;27:3143–3159. doi: 10.1105/tpc.15.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou F. ‘D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling’. Nature. Dec. 2013;504:406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. Dec. 2013;504:401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaux P.-M. Origin of strigolactones in the green lineage. New Phytol. Sep. 2012;195:857–871. doi: 10.1111/j.1469-8137.2012.04209.x. [DOI] [PubMed] [Google Scholar]
- 42.Bythell-Douglas R. Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol. Dec. 2017;15:52. doi: 10.1186/s12915-017-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A. Structure-function analysis of SMAX1 reveals domains that mediate its karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell. May 2020 doi: 10.1105/tpc.19.00752. p. tpc.00752.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates interaction between KAI2 and SMAX1/SMXL, and the degradation of SMAX1 as a result of both MAX2 dependent polyubiquitination and an additional unknown mechanism.
- Wang L. Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell. Apr. 2020;2020 doi: 10.1105/tpc.20.00140. p. tpc.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors found that D14 and KAI2 signalling converged on SMXL2 in controlling hypocotyl length as SMXL2 degradation occurred in response to interaction with D14 or KAI2, depending on the nature of the synthetic ligand.
- 45.Waters M.T., Scaffidi A., Flematti G., Smith S.M. Substrate-induced degradation of the α/β-Fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol Plant. May 2015;8:814–817. doi: 10.1016/j.molp.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 46.Kameoka H., Kyozuka J. Downregulation of rice DWARF 14 LIKE suppress mesocotyl elongation via a strigolactone independent pathway in the dark. J Genet Genom. Mar. 2015;42:119–124. doi: 10.1016/j.jgg.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Zheng J. Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell. Jul. 2020 doi: 10.1105/tpc.20.00123. p. tpc.00123.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Biochemical evidence is provided for D14L, D3, and SMAX1 complex formation in rice. Additionally, TPR2 is the first transcriptional regulator reported to interact with SMAX1 and thus a candidate for the regulation of downstream genes.
- 48.Yoshida S. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. Dec. 2012;196:1208–1216. doi: 10.1111/j.1469-8137.2012.04339.x. [DOI] [PubMed] [Google Scholar]
- Choi J. The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat Commun. Dec. 2020;11:2114. doi: 10.1038/s41467-020-16021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study revealed SMAX1 as a negative regulator of AM symbiosis development and SL biosynthesis in rice roots, thereby demonstrating crosstalk between D14L signalling and SL biosynthesis.
- 50.Gutjahr C. ‘Arbuscular mycorrhiza–specific signaling in rice transcends the common symbiosis signaling pathway’. Plant Cell. Nov. 2008;20:2989–3005. doi: 10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo F. Butenolide derivatives from the plant endophytic fungus Aspergillus terreus. Fitoterapia. Sep. 2016;113:44–50. doi: 10.1016/j.fitote.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Carbonnel S. Lotus japonicus karrikin receptors display divergent ligand-binding specificities and organ-dependent redundancy. PLoS Genet. Dec. 2020;16 doi: 10.1371/journal.pgen.1009249. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article discovered differential responses to synthetic KLs in an organ-specific fashion, pointing towards either differences in the spatial distribution of KL or of KL-transporting or metabolising enzymes.
- 53.Kretzschmar T. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature. Mar. 2012;483:341–344. doi: 10.1038/nature10873. [DOI] [PubMed] [Google Scholar]
- 54.Müller L.M., Harrison M.J. Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. Aug. 2019;50:132–139. doi: 10.1016/j.pbi.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Ito S. Regulation of strigolactone biosynthesis by gibberellin signaling. Plant Physiol. Jun. 2017;174:1250–1259. doi: 10.1104/pp.17.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunsick M. SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat Plants. May 2020 doi: 10.1038/s41477-020-0653-z. [DOI] [PubMed] [Google Scholar]; This study discovered that supressed seed germination can be overridden by SL-induced activation of D14L signalling and removal of SMAX1 in Arabidopsis expressing members of the S. hermonthica D14L (HTL) receptors.
- 57.Foo E. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol Plant. 2013;6:76–87. doi: 10.1093/mp/sss115. [DOI] [PubMed] [Google Scholar]
- 58.Gutjahr C. The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice: Role of rice STR1 and STR2 in arbuscule formation. Plant J. 2012;69:906–920. doi: 10.1111/j.1365-313X.2011.04842.x. [DOI] [PubMed] [Google Scholar]
- 59.Gomez-Roldan V. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 60.Vogel J. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato: SlCCD7 controls tomato branching. Plant J. 2010;61:300–311. doi: 10.1111/j.1365-313X.2009.04056.x. [DOI] [PubMed] [Google Scholar]
- 61.Kohlen W. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8(SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012;196:535–547. doi: 10.1111/j.1469-8137.2012.04265.x. [DOI] [PubMed] [Google Scholar]
- 62.Yu N. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2014;24:130–133. doi: 10.1038/cr.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Floss D. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci. 2013;110:E5025–E5034. doi: 10.1073/pnas.1308973110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foo E. Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann Bot. 2013;111:769–779. doi: 10.1093/aob/mct041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martín-Rodríguez J.A. Role of gibberellins during arbuscular mycorrhizal formation in tomato: new insights revealed by endogenous quantification and genetic analysis of their metabolism in mycorrhizal roots. Physiol Plant. 2015;154:66–81. doi: 10.1111/ppl.12274. [DOI] [PubMed] [Google Scholar]