Figure 1.
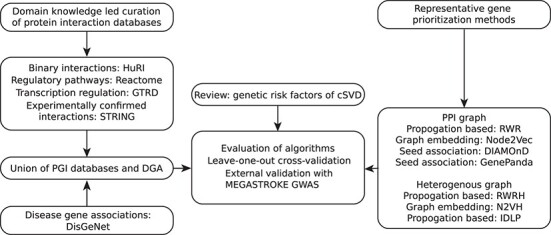
Benchmarking workflow of network-based gene prioritization in cSVD. There are three main components of the benchmarking workflow: assembling input networks, selection of algorithms and validation of algorithms. (i) PGI was assembled with domain knowledge-lead curation of protein/gene interactions from four databases. DGAs were added to PGI to generate disease–gene heterogeneous networks. (ii) Representative gene prioritization algorithms were selected based on the originality with the non-network-based algorithms or hybrid algorithms excluded. (iii) Performance of algorithms was assessed with LOOCV and externally validated with MEGASTROKE GWAS results. Abbreviations: PGI, protein–gene interaction; DGA, disease–gene association; cSVD, cerebral small vessel disease; LOOCV, leave-one-out cross-validation; GWAS, genome-wide association study. Please see Table 1 for full names of databases and Table 2 for full names of algorithms.
