Figure 4 .
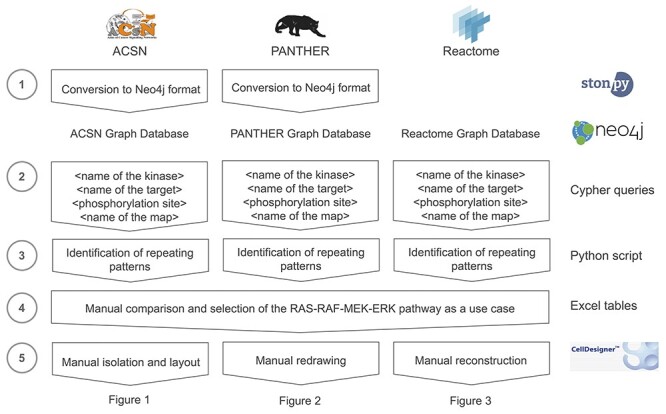
The workflow for identifying repeated patterns in the ACSN, PANTHER and Reactome databases. The steps include: (1) storing processes from the databases in the Neo4j format; (2) querying phosphorylation processes and representing them in the form of quadruplets; (3) organizing repeated phosphorylation processes according to their occurrence in maps; (4) manually revising and finding repeated processes differently represented in maps and selecting the use case; (5) manually isolating the RAS–RAF–MEK–ERK pathway variants for making them visually comparable.
