Figure 1 .
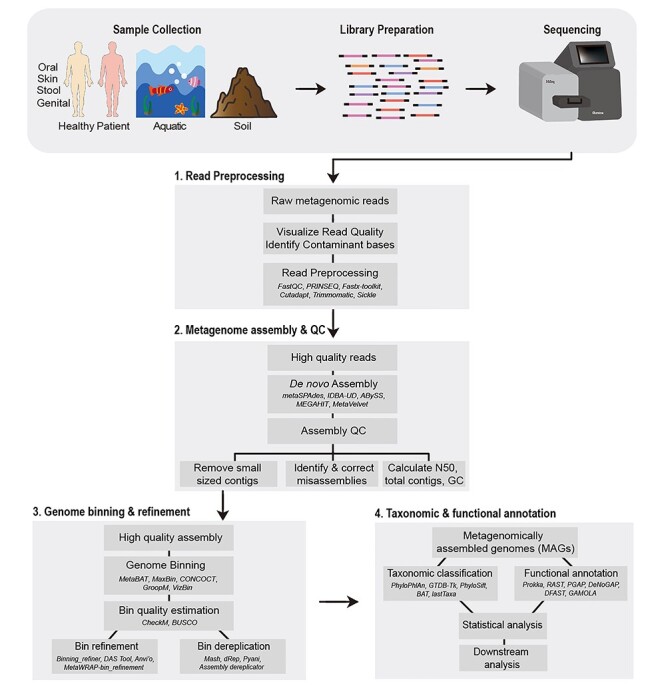
Schematic representation of the workflow for genome-resolved metagenomics analysis. The typical genome-resolved analysis of metagenomes, obtained through any source, typically involves four steps i.e. (1) read preprocessing, (2) metagenome assembly and QC, (3) genome binning and refinement and (4) taxonomic and functional annotation of the recovered MAGs. In the figure, examples of several tools for each of the steps is also provided (names italicized).
