FIGURE 1.
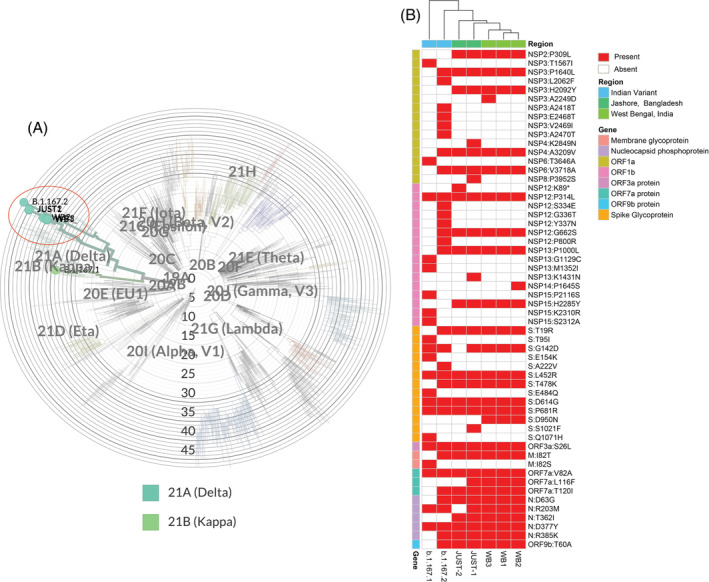
Comparison of amino acid substitutions retrieved from genome sequences of seven SARS‐CoV‐2 viruses. A, Phylogenetic tree of SARS‐CoV‐2 uses five isolates (JUST1, JUST2, WB1, WB2, WB3 and two references [B.1.167.1 and B.1.167.2 strains]). The study isolates were highlighted with circle. The tree was condensed using Nextclade (https://clades.nextstrain.org/). B, Amino acid substitutions found in our study samples (JUST1, JUST2) are compared with three West Bengal (WB1, WB2, WB3) and reference B.1.167.1 and reference B.1.167.2 strains. Mutations and amino acid substitutions were retrieved from Nextclade (https://clades.nextstrain.org/)
