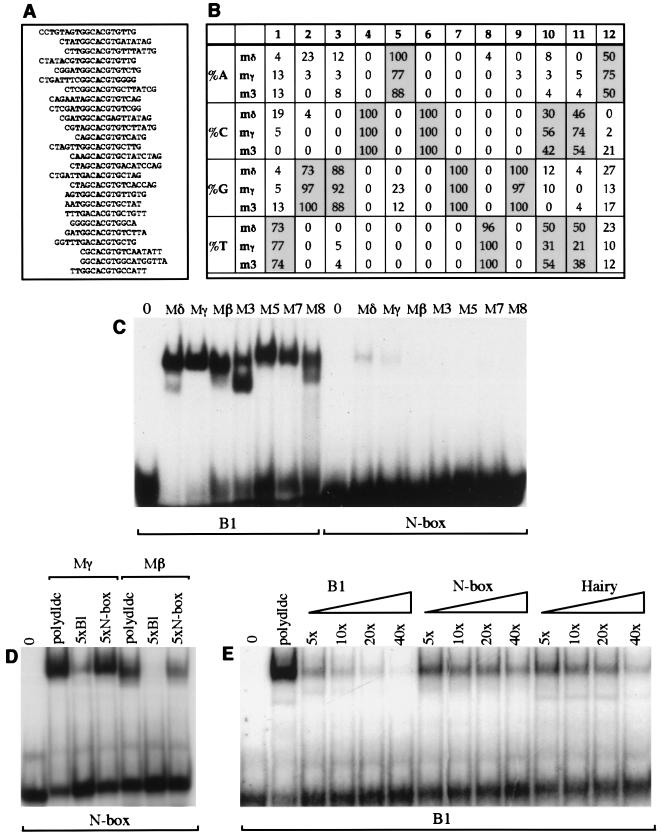
FIG. 1.
Identification of optimal targets for E(spl)bHLH proteins. (A and B) Binding site selection was used to identify optimal E(spl)bHLH DNA targets. (A) Alignment of 26 sequences obtained after five rounds of selection with the Mδ protein. Selection of oligonucleotides with Mδ was performed in two separate experiments; soluble Mδ protein extract was used to select the first 17 oligonucleotides listed. The soluble and refolded Mδ extracts show no obvious differences in DNA binding preferences. (B) The nucleotides present at each position of fifth-round sequences selected by the Mδ (n = 26), Mγ (n = 39), and M3 (n = 24) proteins (n is the number of sequences analyzed). Nucleotides selected with a frequency of 50% or higher are highlighted. (C) Binding of all seven E(spl)bHLH proteins to the optimal E(spl) consensus (B1; contains a palindromic version of the ESE box) or the N-box oligonucleotide. Identical amounts of protein (∼50 ng) and labeled oligonucleotides were used for the equivalent reactions. (D) Addition of a fivefold molar excess of unlabeled B1 oligonucleotide has a greater effect on binding of the Mγ and Mβ proteins (approximately 500 ng of soluble pGex fusion protein) to the N-box probe than addition of a fivefold excess molar excess of unlabeled N-box oligonucleotide. (E) Binding of Mγ and Mβ proteins (approximately 50 ng of pGex fusion protein in a soluble bacterial extract) to labeled B1 probe in the presence of nonspecific competitor DNA [poly(dI-dC)] or 5-, 10-, 20-, or 40-fold molar excess of unlabeled B1, N-box, or Hairy oligonucleotide as indicated. Sequences of all oligonucleotides used are listed in Table 1. The lanes 0 in panels C to E contain labeled oligonucleotides in the absence of added protein.