FIG. 3.
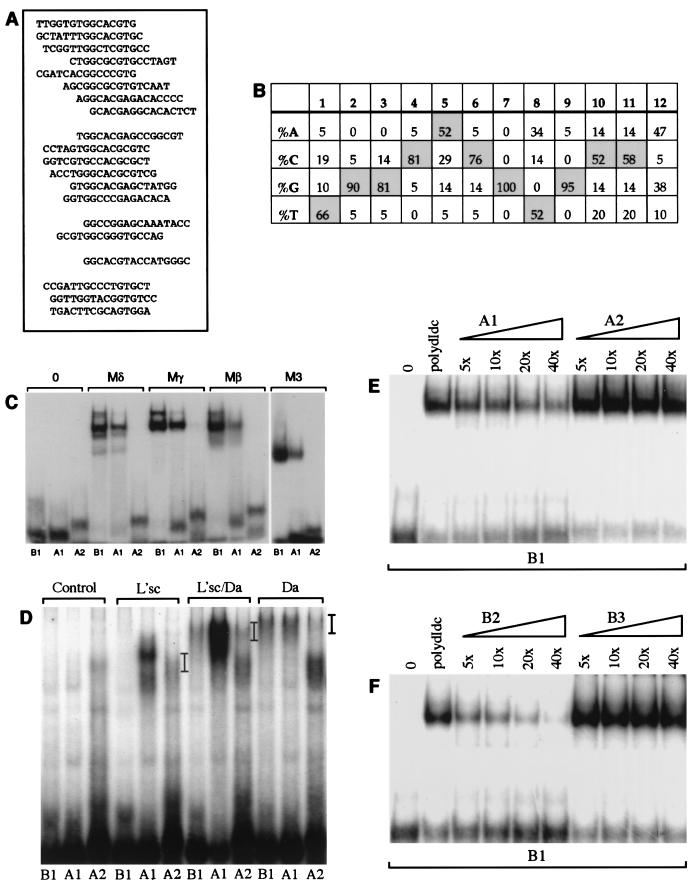
Sequences flanking the E-box core influence binding of E(spl)bHLH and proneural proteins. (A and B) Sequences selected after three rounds of binding with the Mδ protein are listed (A) and summarized to show the frequency of nucleotides at each position (B). The top 15 oligonucleotides listed contain a CNCGNG core, followed by 2 with suboptimal bases at position 6 in the E box and 1 with a suboptimal base at position 9. The final three oligonucleotides contain GTG, half of a class B E-box, but may not bind the E(spl)bHLH proteins with high affinity. (B) After three rounds of selection, there is a strong preference for certain nucleotides in the positions flanking the E box (a similar preference is evident if the analysis is restricted to the 15 oligonucleotides containing a CNCGNG core [data not shown]). (C to F) Effect of flanking sequence on DNA binding in vitro. (C and D) Binding of E(spl)bHLH proteins to B1, A1, and A2 (Table 1). The B1 and A1 sites contain optimal flanking bases and differ by a single-base substitution that switches the E box from a class B site (B1) to a class A site (A1) (10). The A2 site has the class A E-box core but suboptimal flanking sequences. (C) Binding of Mδ, Mβ, Mγ, and M3 proteins (75 ng) can be detected with the B1 and A1 sites but not with A2. (D) Binding of proneural proteins to the different E-box oligonucleotides (as indicated) was tested by using L’sc protein extract only, a 1:1 mixture of L’sc and Da extracts, and Da extracts only. Position of the DNA-protein complexes are indicated by bars. Negative control reactions containing soluble bacterial extract (Control) were included for comparison. (E) Effects of 5-, 10-, 20-, or 40-fold molar excess of either unlabeled A1 or A2 oligonucleotide on binding of Mγ (50 ng) to the B1 probe. Addition of unlabeled A1 diminishes binding to B1 in a concentration-dependent manner; 40-fold molar excess of unlabeled A2 did not compete with the B1 probe (confirmed by phosphorimaging analysis). (F) Mγ binding to the B1 probe in the presence of 5-, 10-, 20-, or 40-fold molar excess of either unlabeled B2 and B3 oligonucleotides. B2 differs from B1 by two suboptimal nucleotides in the flanking sequences, and B3 has the least optimal bases at all positions flanking the E-box core (using the information from Fig. 1B). B2 competes with B1 in a concentration-dependent manner, although not as efficiently as B1 (Fig. 1B), while 40-fold molar excess of unlabeled B3 did not compete with the B1 probe (confirmed by phosphorimaging). The amounts of protein and probe used for panels E and F were identical to the amounts used for Fig. 1E. The sequences of the oligonucleotides used in panels C to F are listed in Table 1. Lanes 0 contain labeled oligonucleotides in the absence of added protein.