Figure 3.
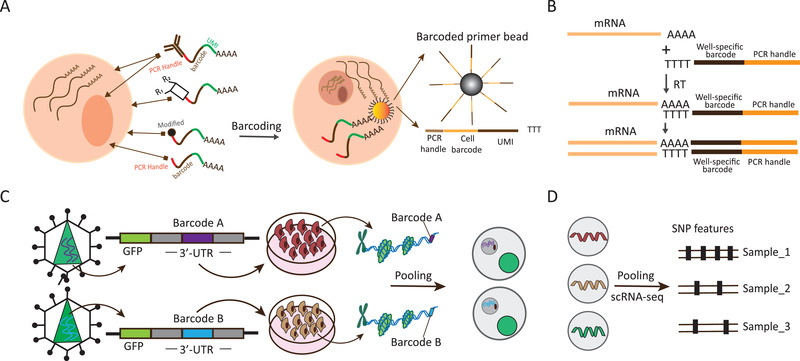
Schematic overview of four DNA‐based barcoding strategies. A) S1: Oligo‐dA‐based barcoding. For this strategy, sample‐specific DNA barcode is generally polyadenylated at the 3′‐end and is structurally similar to endogenous mRNA. It can be captured alongside mRNA by the same barcoded bead that is cell‐specific. Several direct and indirect approaches are available for assigning sample barcodes to cells, depending on sample type, for example, polyadenylated barcoded ssDNA, which diffuses into a fixed nucleus and directly labels the mRNA from nucleus; chemically modified barcode oligonucleotides, which bind with cellular proteins for cell tagging; and a transient transfection with a barcode, which labels the cell. As an indirect labeling approach, barcoded antibody and lipid‐tagged barcode are used to target ubiquitous proteins or the nucleus core complex, and the cellular membrane, accordingly, to label the cell. B) S2: Multiplexing by integration of DNA barcode with mRNA. The integrated cDNA is generated via a reverse‐transcription (RT) reaction with sample‐specific barcoded primers that are poly(dT) at the 3′‐end or have the sequences complementary to transcript. C) S3: Viral integration‐based genetic barcoding. Sample barcodes (barcode A, barcode B) and green fluorescent protein (GFP) are merged to be incorporated into genome sequence respectively via lentiviral transduction. Sample barcode can be transcribed into polyadenylated transcripts, which are efficiently captured along with endogenous transcripts in single‐cell library construction. D) S4: Exploitation of naturally occurring genetic mutations. Here, natural mutations are used as barcodes of individuals or cancer cell lines. Demultiplexing is mainly based on the dissection of SNP variation using computational tools, such as demuxlet.
