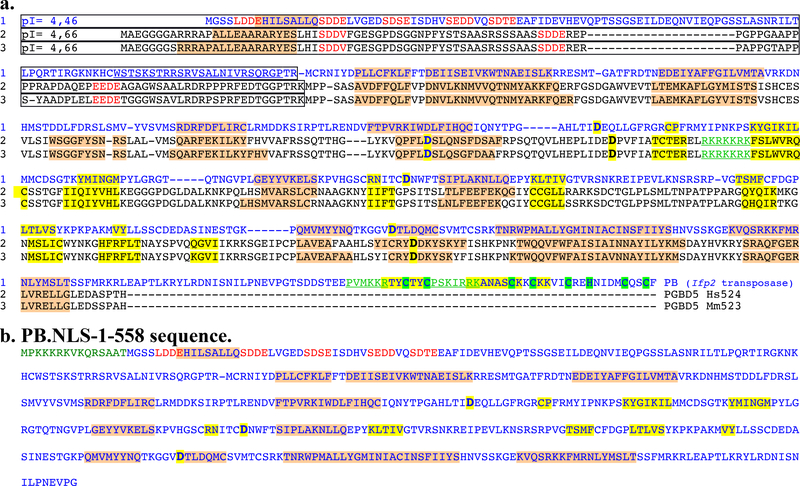
Fig. 4. Sequence features of the Ifp2 transposase (PB) variants and two Mm523-like PGBD5 isoforms.
(a) Protein sequence alignment of Ifp2 transposase (PB) with two murine and human domesticated PGBD5 proteins corresponding to the orthologous Hs524 and Mm523 isoforms. (b) Sequence features of PB.NLS-1–558. Secondary structure predictions calculated with psipred (http://bioinf.cs.ucl.ac.uk/psipred/) and Jpred4 (http://www.compbio.dundee.ac.uk/jpred/) were highlighted in pink for α-helices and in orange for β-strands. The three proteins share two domains: a N-terminal domain that was few structured, with an acid pI (boxed regions) and repeated acid motifs (in red letters), a domain of ~400 amino acid residues that display a basic pI. PB contained a third C-terminal domain, the CRD, that contains cysteins (highlighted in green) able to assemble zinc finger folds. Aspartic residues inactivating the recombinase catalytic activity were bolded and highlighted in yellow [17,21]. The PB NLS and the putative NLS in PGBD5 isoforms were underlined and typed in green.