Abstract
Aeromonas salmonicida subsp. salmonicida, the causative agent of furunculosis, has extensive negative effects on wild and farmed salmonids worldwide. Vaccination induces some protection under certain conditions but disease outbreaks occur even in vaccinated fish. Therefore, alternative disease control approaches are required to ensure the sustainable expansion of rainbow trout aquaculture. Selective breeding can be applied to enhance host resistance to pathogens. The present work used genome-wide association study (GWAS) to identify quantitative trait loci (QTL) associated with A. salmonicida resistance in rainbow trout. A total 798 rainbow trout exposed to A. salmonicida by bath challenge revealed 614 susceptible and 138 resistant fish. Genotyping was conducted using the 57 K single nucleotide polymorphism (SNP) array and the GWAS was performed for survival and time to death phenotypes. We identified a QTL on chromosome 16 and located positional candidate genes in the proximity of the most significant SNPs. In addition, samples from exposed fish were examined for expression of 24 immune-relevant genes indicating a systematic immune response to the infection. The present work demonstrated that resistance to A. salmonicida is moderately heritable with oligogenic architecture. These result will be useful for the future breeding programs for improving the natural resistance of rainbow trout against furunculosis.
Subject terms: Gene expression analysis, Genome-wide association studies, Bacterial infection, Gene regulation in immune cells
Introduction
Aeromonas salmonicida subsp. salmonicida is a ubiquitous Gram-negative bacterium, causing furunculosis in wild and captive salmonids in fresh- and saltwater worldwide1. Its high pathogenic potential is a serious threat in aquaculture, causing significant morbidity and mortality in intensive salmonid rearing systems2. Elevated water temperatures are beneficial for the growth of A. salmonicida and high stocking densities in aquaculture increase transmission of bacteria between fish3. Climate changes may increase the prevalence of furunculosis in both wild4 and aquacultured fish5, and with the expanding aquaculture activities and increasing water temperatures no self-limitations are at hand.
Fish vaccines are available and confer some protection as the injection of mineral oil-adjuvanted bacterin vaccines induces a strong immune reaction reflected by high serum antibody levels6,7. However, furunculosis is still a serious threat to Danish maricultured rainbow trout (Oncorhynchus mykiss) and disease outbreaks occur even among vaccinated fish during summer periods with higher water temperatures8. Vaccination procedures may lead to marked production losses due to the starvation of fish before and after vaccination and the use of antibiotic treatments should be avoided due to environmental concerns. Therefore, alternative disease control approaches are required to ensure the sustainable expansion of rainbow trout aquaculture.
Selective breeding programs for disease resistant traits have a considerable potential9. For example, genetic resistance against infectious pancreatic necrosis virus (IPNV) was successfully improved in Atlantic salmon (Salmo salar) based on identification of a major QTL10,11. A major QTL in rainbow trout was found associated with resistance to Vibrio anguillarum12 and Flavobacterium psychrophilum13. QTL were linked to resistance against parasitic diseases such as amoebic gill disease (AGD) in Atlantic salmon14 and ciliate Ichthyophthirius multifiliis in rainbow trout15.
Attempts to identify brook trout (Salvenius fontinalis) strains resistant to A. salmonicida have been ongoing almost for a century16–20 showing a basis for selection programs based on significant genetic variation. This was also found for Atlantic salmon21,22 and the higher survival rates were later associated with MHC allele variation23,24. The protective mechanism was suggested to be elevated serum hemolytic activity in salmon25 and higher serum bactericidal activity in rainbow trout26,27.
GWAS has shown to be a promising tool to improve the genetic status of fish used for aquaculture purposes and the 57 K SNP panel28 has previously disclosed the genetic architecture of several traits of interest in rainbow trout12,15,29–35. Therefore the aim of this study was to assess the prospective of using GWAS to identify potential QTL for disease resistance against A. salmonicida. We genotyped 752 rainbow trout exposed to A. salmonicida through bath challenge during 13 days using the 57 K SNP panel and performed the GWAS for survival and time to death (TD) phenotypes to reveal QTL associated to A. salmonicida resistance. To gain insight of underlying mechanisms of resistance, we identified the positional candidate genes in the proximity of the QTL from the most recent assmbly of rainbow trout reference genome (USDA_OmykA_1.1 (GCF_013265735.2, NCBI)) and compared the expression profiles of immune-relevant genes in different fish groups.
Results
Mortality
The first moribund fish was recorded at 1 dpc (Fig. 1). From there on, the mortality increased sharply until 6 dpc (76%) and only few mortalities occurred thereafter. Mortality varied between the tanks to some extent (Supplementary Table S1) and the total mortality of 82% was recorded at 13 dpc.
Figure 1.
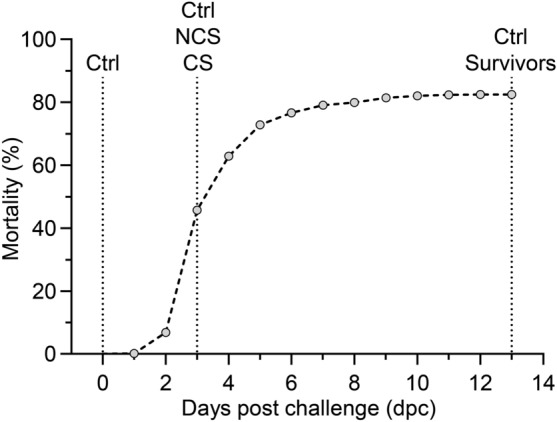
Kaplan–Meier plot (constructed with GraphPad Prism software) showing the development of mortality in 798 rainbow trout after waterborne infection with Aeromonas salmonicida subsp. salmonicida strain 111129–1/2. The curve represents cumulative mortality (%) of fish in 12 tanks. Dotted vertical lines mark the sampling time points for qPCR (Ctrl- non-exposed control fish, NCS—exposed fish with no clinical signs, CS—exposed fish with clinical signs, Survivors—exposed fish surviving the challenge).
Genetic analysis
The results from REML analysis are presented in Table 1 and show that heritability to furunculosis infection was relatively high for survival (22% on the observed scale, 47% on the underlying scale of liability) and TD (36% on the observed scale). The estimated variance for additive genetic effects were 0.033 ± 0.010 and 4.975 ± 1.126 for survival and TD, respectively.
Table 1.
Estimated variance of components from REML analysis of survival and time to death (TD) using a genomic animal model.
| Factor | Survival | TD |
|---|---|---|
| Additive genetic variance | 0.033 ± 0.010 | 4.975 ± 1.126 |
| Residual variance | 0.116 ± 0.008 | 9.031 ± 0.657 |
| Phenotypic variance | 0.149 ± 0.009 | 14.007 ± 0.967 |
| Heritability (observed scale) | 0.222 ± 0.059 | 0.355 ± 0.062 |
| Heritability (underlying scale) | 0.471 ± 0.125 | – |
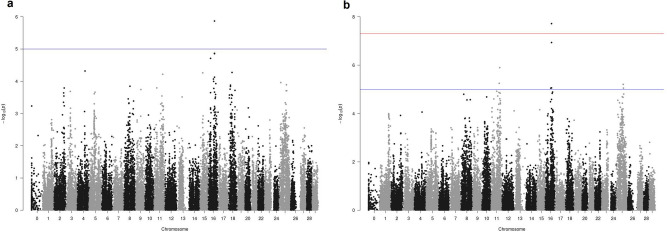
The Manhattan plots for survival and TD (Fig. 2), as results of the LOCO-GWAS, indicate a significant QTL on chromosome 16, explaining 17% of the genetic variance for TD (Table 2). The same SNP (AX-89969631) came up as the most significant for both survival and TD, however, it was only clearly significant for TD (1.91e-8), likely due to a higher heritability for this trait.
Figure 2.
The Manhattan plot from LMM-LOCO GWAS of survival (a) and time to death (DT) (b) in rainbow trout after exposure to A. salmonicida subsp. salmonicida. The y-axis shows the p value for each SNP and x-axis individual SNPs. The red horizontal line represent the genome-wide significance threshold of p-value 5e−08 and the blue horizontal line indicates the suggestive level of significance with p-value of 1e−05.
Table 2.
The top SNPs for TD and survival.
| Chr | SNP | Position | Type | Freq | Effect | SE | P | Fr. genvar | Fr. phenvar | |
|---|---|---|---|---|---|---|---|---|---|---|
| USDA_OmykA_1.1 (GCF_013265735.2) | Omyk_1.0 (GCF_002163495.1) | |||||||||
| Time to death (DT) | ||||||||||
| 16 | AX-89969631 | 48210249 | 44822522 | C/T | 0.84 | − 1.76 | 0.31 | 1.91e−08 | 17% | 6% |
| 16 | AX-89973999 | 48231736 | 44846688 | G/T | 0.80 | − 1.49 | 0.28 | 1.16e−07 | 14% | 5% |
| 11 | AX-89976064 | 65121697 | 61934614 | C/A | 0.25 | − 1.34 | 0.28 | 1.26e−06 | 14% | 5% |
| 11 | AX-89919380 | 61196699 | 58124529 | C/T | 0.15 | − 1.42 | 0.31 | 5.66e−06 | 10% | 4% |
| 25 | AX-89933102 | 10230926 | 49243042 | C/T | 0.55 | − 1.04 | 0.23 | 6.25e−06 | 11% | 4% |
| 16 | AX-89924175 | 48220966 | 44834124 | G/A | 0.68 | − 1.16 | 0.26 | 8.72e−06 | 12% | 4% |
| 25 | AX-89956080 | 11916756 | 50865587 | C/A | 0.41 | 1.06 | 0.24 | 8.92e−06 | 11% | 4% |
| 16 | AX-89926977 | 42909875 | 39677850 | G/A | 0.09 | 1.69 | 0.38 | 8.96e−06 | 9% | 3% |
| 11 | AX-89965839 | 42857280 | 39901842 | G/T | 0.22 | − 1.42 | 0.32 | 1.20e−05 | 14% | 5% |
| 16 | AX-89934677 | 56176558 | 52680845 | C/T | 0.52 | 1.30 | 0.30 | 1.30e−05 | 17% | 6% |
| 11 | AX-89942657 | 59019488 | 56061980 | C/A | 0.84 | 1.33 | 0.31 | 1.35e−05 | 10% | 3% |
| 11 | AX-89963218 | 67994309 | 64707318 | C/A | 0.88 | 1.47 | 0.34 | 1.44e−05 | 9% | 3% |
| 16 | AX-89919325 | 53039122 | 49570500 | C/T | 0.43 | − 1.10 | 0.25 | 1.47e−05 | 12% | 4% |
| 25 | AX-89957036 | 13953432 | 52861958 | C/T | 0.50 | − 0.88 | 0.20 | 1.57e−05 | 8% | 3% |
| Survival | ||||||||||
| 16 | AX-89969631 | 48210249 | 44822522 | C/T | 0.84 | − 0.16 | 0.033 | 1.35e−06 | 20% | 5% |
| 16 | AX-89924175 | 48220966 | 44834124 | G/A | 0.68 | − 0.12 | 0.03 | 1.36e−05 | 19% | 4% |
| 16 | AX-89973999 | 48231736 | 44846688 | G/T | 0.80 | − 0.13 | 0.03 | 1.39e−05 | 16% | 4% |
| 16 | AX-89976723 | 19107648 | 16278865 | C/A | 0.55 | − 0.10 | 0.02 | 1.95e−05 | 14% | 3% |
| 4 | AX-89965612 | 12013837 | 54733880 | G/A | 0.14 | 0.16 | 0.04 | 4.81e−05 | 19% | 4% |
| 15 | AX-89918838 | 22601764 | 19112526 | G/A | 0.44 | 0.11 | 0.03 | 5.48e−05 | 18% | 4% |
| 11 | AX-89976064 | 65121697 | 61934614 | C/A | 0.25 | − 0.12 | 0.03 | 6.07e−05 | 16% | 3% |
| 16 | AX-89964054 | 48349717 | 44952397 | C/A | 0.47 | − 0.11 | 0.03 | 7.48e−05 | 18% | 4% |
| 16 | AX-89955003 | 40411668 | 37206251 | C/A | 0.90 | − 0.15 | 0.04 | 8.53e−05 | 14% | 3% |
The two last columns give the estimated fraction of genetic and phenotypic variance explained by each SNP. SNPs are listed based on significance.
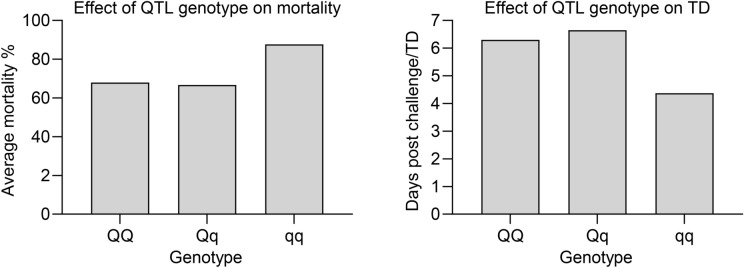
The calculation of the average observed mortality for fish having different genotypes for the most significant SNP is presented in Fig. 3. The favorable homozygote (QQ) and heterozygote (Qq) had an overall mortality of 66% while the mortality for the unfavorable homozygote (qq) was 88%. A similar tendency was observed for TD.
Figure 3.
Average observed mortality for different genotypes: (a) survival and (b) time to death (TD) after challenge with A. salmonicida subsp. salmonicida for the top SNP (AX-89969631) at chromosome 16.
Candidate genes
Positional candidate genes within the 1000 Kb window (47 750 K–48 750 K) of the most significant SNPs (AX-89969631 and AX-89973999) associated with resistance on chromosome 16 (NC_048580.1) was identified using the latest assembly of rainbow trout genome USDA_OmykA_1.1 (GCF_013265735.2) in NCBI-Genbank. Among the 33 genes located in this region, we found 30 protein-coding genes, 1 pseudogene and 2 non-coding genes (Table 3). The most significant SNP (AX-89969631) was located in the LOC118939552 region of non-coding RNA while the gene encoding PRA1 family protein 3 mapped into the second most significant SNP (AX-89973999).
Table 3.
Positional candidate genes within the 1000 Kb window (47,750 K–48,750 K) of the two most significant SNPs (AX-89969631 and AX-89973999) from the latest assembly of the rainbow trout genome USDA_OmykA_1.1 (GCF_013265735.2) in NCBI-Genbank.
| Gene ID | Description | Type | Location | Strand |
|---|---|---|---|---|
| LOC110492148 | Calmodulin-binding transcription activator 1 | Protein coding | 47409235–47908274 | − |
| LOC110492150 | dnaJ homolog subfamily C member 11 | Protein coding | 47918157–47927478 | − |
| thap3 | THAP domain containing, apoptosis associated protein 3 | Protein coding | 47918157–47927478 | − |
| LOC110492151 | F-box only protein 6 | Protein coding | 47932110–47950665 | + |
| LOC110492153 | Cell division control protein 42 homolog | PROTEIN coding | 47953418–47956955 | − |
| LOC118939548 | Uncharacterized | ncRNA | 47981859–47988731 | + |
| LOC110492154 | Lamina-associated polypeptide 2, isoforms beta/delta/epsilon/gamma | Protein coding | 48006228–48010038 | + |
| LOC118939547 | N-fatty-acyl-amino acid synthase/hydrolase PM20D1.2-like | Protein coding | 48010169–48026769 | + |
| LOC118939549 | Acidic mammalian chitinase-like | Protein coding | 48031902–48036060 | − |
| LOC118939550 | Neurofascin-like | Pseudo | 48042033–48074973 | − |
| LOC118939551 | Calcium-independent phospholipase A2-gamma-like | Protein coding | 48076504–48087589 | − |
| LOC110492156 | Uncharacterized protein At5g50100, chloroplastic | Protein coding | 48089427–48093940 | − |
| LOC110491082 | Monocarboxylate transporter 2-like | Protein coding | 48101686–48113512 | + |
| LOC110492157 | Chemokine-like protein TAFA-1 | Protein coding | 48145652–48171888 | + |
| LOC110491083 | chemokine-like protein TAFA-4 | Protein coding | 48184977–48196697 | − |
| LOC118939552 | Uncharacterized LOC118939552 | ncRNA | 48208293–48212220 | − |
| LOC110492159 | NEDD8-activating enzyme E1 catalytic subunit | Protein coding | 48220758–48230211 | − |
| LOC110492162 | PRA1 family protein 3 | Protein coding | 48231344–48235518 | + |
| LOC110492161 | Leiomodin-3 | Protein coding | 48234990–48241496 | − |
| LOC110492160 | FERM domain-containing protein 4B | Protein coding | 48242058–48272289 | − |
| LOC110492164 | Microphthalmia-associated transcription factor | Protein coding | 48282132–48331926 | + |
| LOC110492165 | 2-epi-5-epi-valiolone synthase | Protein coding | 48333920–48343017 | − |
| LOC110492167 | myoD family inhibitor domain-containing protein 2-like | Protein coding | 48343132–48347793 | − |
| LOC110492168 | forkhead box protein P1-B | Protein coding | 48348467–48371945 | − |
| LOC110492170 | Eukaryotic translation initiation factor 4E type 3 | Protein coding | 48372037–48383460 | − |
| LOC110492169 | Serine/threonine-protein phosphatase 4 regulatory subunit 2-A | Protein coding | 48383542–48393271 | + |
| LOC110491084 | Guanylyl cyclase-activating protein 2 | Protein coding | 48400096–48402355 | + |
| oard1 | O-acyl-ADP-ribose deacylase 1 | Protein coding | 48403279–48405646 | + |
| LOC110492174 | Synaptotagmin-2 | Protein coding | 48407064–48483333 | + |
| LOC110492173 | Protein phosphatase 1 regulatory subunit 12B | Protein coding | 48483470–48521507 | − |
| LOC110492176 | Neurofilament medium polypeptide | Protein coding | 48525042–48533176 | − |
| LOC110491085 | Immunoglobulin-like and fibronectin Type III domain-containing protein 1 | Protein coding | 48547180–48548167 | − |
| LOC110492177 | Metabotropic glutamate receptor 4 | Protein coding | 48650173–48970488 | − |
The genes highlighted in bold include the two most significant SNPs.
Immune gene expression
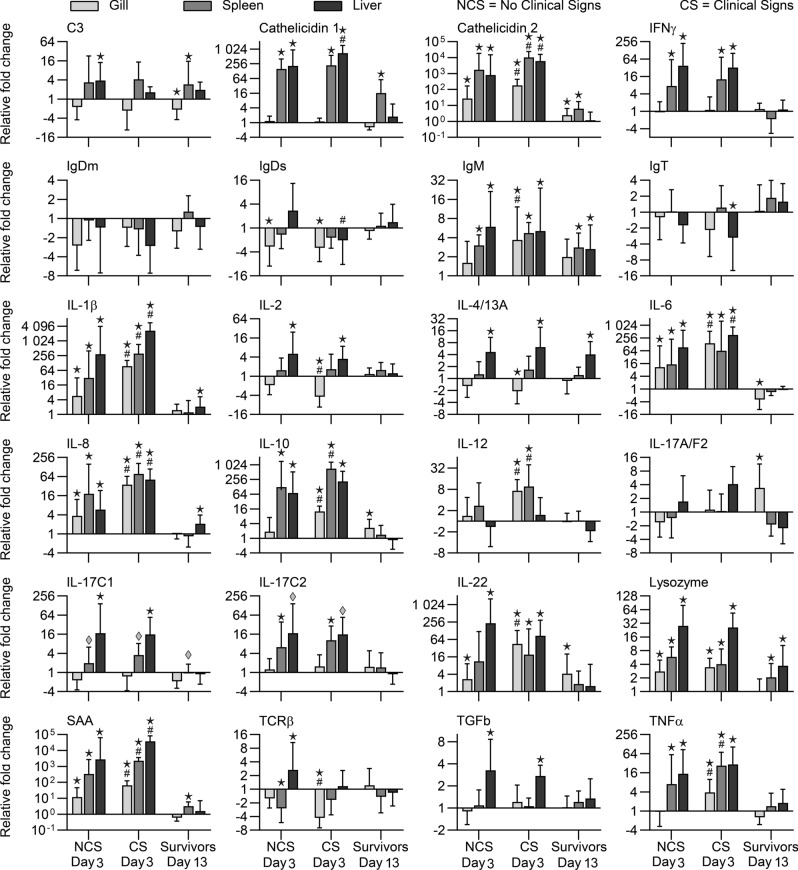
Significantly expressed immune-relevant genes are displayed in Fig. 4 reflecting different involvement of three different organs in exposed fish when compared to non-exposed controls. The majority of genes were highly up-regulated in the liver, while less regulated genes were recorded in the gills. The majority of the significant regulations occurred at 3 dpc in the fish exposed to bacteria and the highest regulations were found in the fish showing clinical signs (CS). The A. salmonicida exposed fish that were not showing clinical signs (NCS) exhibited similar regulation of the same genes, although to a lower degree.
Figure 4.
Different expression patterns of cytokine and effector molecule genes after infection with A. salmonicida subsp. salmonicida in rainbow trout. NCS: exposed fish with no clinical signs, CS: exposed fish with clinical signs, Survivors—exposed fish surviving the challenge at 13 dpc. Star: Significant differences (p < 0.05) between non-exposed (control) groups and exposed groups—NCS, CS and survivors. #Significant differences (p < 0.05) between the NCS and CS groups at 3 dpc. Diamond: Significant differences (p < 0.05) with non-parametric Mann–Whitney test were applied for fish groups that had less than 3 positive Cq values.
Expression at 3 dpc
All the genes except those encoding IgT, IgDs and IgDm were significantly regulated in at least one of the sampled organs of infected fish at 3 dpc.
Liver
Genes encoding innate immune effector molecules (C3, cathelicidins, lysozyme, SAA) were significantly up-regulated in the liver of infected fish. Up-regulation of the gene encoding IgM was recorded whereas other immunoglobulin genes did not show a clear involvement. A substantial significant up-regulation occurred for the genes for the main pro-inflammatory cytokines (IL-1β, IL-6, IL-8, TNFα, IFNγ), regulatory cytokines (IL-10, TGFβ) and cytokines involved in functions of different Th cell subsets (IL-2, IL-3/14a, IL-17C1, IL-17C2, IL-22, TCRβ).
Spleen
The majority of genes in the spleen were generally expressed at a lower level when compared to the liver, but the genes encoding cathelicidin 2, IL-8, IL-10 and IL-12 showed a higher-fold splenic up-regulation. Genes encoding IL-4/13a, IL-17A7/F2, IL-22, TGFβ and C3 showed no significant regulation in the spleen.
Gills
The gene expression in the gills was primarily seen in fish with clinical signs. In some cases, we found that genes significantly up-regulated in one or both of the other organs, were significantly down-regulated in gills (IL-2, IL-4/13a, C3) or showed no significant regulation (IL-17A/F2, IL-17C1, IL-17C2, TGFβ, IFNγ, cathelicidin 1). Cytokine genes (IL-1β, IL-6, IL-8, TNFα, IL-10, IL-12, IL-22) and innate immune genes encoding for cathelicidin 2, lysozyme and SAA were in most cases significantly up-regulated in infected groups. Genes encoding immunoglobulins were mainly down-regulated, with the exception of a significant up-regulation for IgM in the gills of fish showing clinical signs.
Expression at 13 dpc
In the late phase of infection, marked by a flattened mortality curve, surviving fish showed a lower gene expression level.
Liver
Genes encoding IL-1β, IL-4/13a, IL-8, lysozyme and IgM were moderately up-regulated in surviving fish.
Spleen
Genes encoding C3, cathelicidins, lysozyme, SAA, IgM and IL-17C1 were significantly up-regulated at this late infection time-point.
Gills
The gene encoding C3 was slightly but significantly down-regulated, and the gene for cathelicidin 2 was up-regulated in fish surviving the infection. A slight significant up-regulation occurred also for the cytokine genes encoding IL-10, IL-17A/F2 and IL-22, while the gene for IL-6 was down-regulated.
Bacterial load
Bacterial infection levels at 3 dpc are illustrated in Fig. 5. Fish with clinical signs had higher level of bacterial transcripts compared to the fish that were exposed to bacteria but had no clinical signs. In general, only few fish without clinical signs had bacterial transcripts in their organs (1/15 and 2/15 for liver and spleen, respectively). No bacterial transcripts were found in the organs of surviving fish at 13 dpc.
Figure 5.
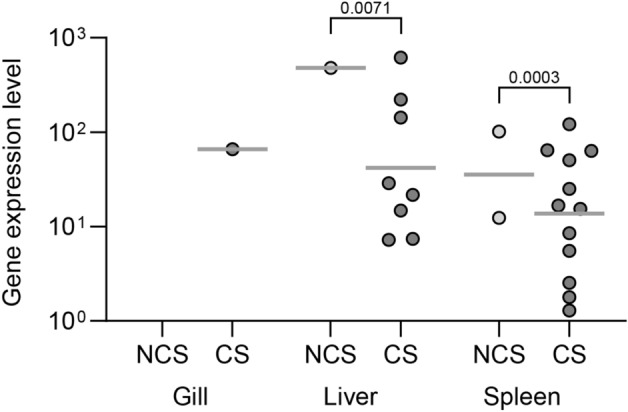
Level of A. salmonicida subsp. salmonicida gene (aopO) transcripts in different organs of fish at 3 days post challenge (dpc): NCS: exposed fish with no clinical signs, CS: exposed fish with clinical signs. Fifteen fish from each group were investigated. Number above the square brackets indicate p values between groups. For the NCS group, only few fish showed positive Cq values, therefore qualitative assessment was applied (presence or absence of Cq values) and data was analyzed with nonparametric Mann–Whitney test.
Discussion
Furunculosis is among the main bacterial diseases affecting rainbow trout in Danish marine aquaculture farms. Vaccination has shown to protect salmonids under certain conditions6 but disease outbreaks among vaccinated fish often occur in Danish rainbow trout mariculture farms when water temperatures exceed 20 °C8. This calls for novel approaches of disease prevention and in this context, genetic breeding of disease resistant rainbow trout may be a possible way forward. In the present study we searched for a QTL associated with furunculosis resistance in rainbow trout applicable for new breeding programs. In addition, we aimed to detect the genetic regions and genes that explain the resistance and supported this approach with immune gene expression studies of susceptible and resistant trout. The genome-wide association study involving 752 rainbow trout exposed to A. salmonicida showed that heritability of furunculosis resistance in trout was relatively high, both regarding survival and time to death phenotypes. Mortality after the bath challenge was high (82%), but declined and stabilized towards the end of the challenge period. This indicated that the survivors are not just fish with longer incubation period, but truly resistant individuals. This notion should, however, be further elucidated by increasing samplings during the entire study period (and confirm absence of bacteria in survivors) and by re-exposing survivors to A. salmonicida to confirm true resistance. A significant QTL was found on chromosome 16 carrying SNPs being significant for both survival and TD. However the most significant SNP reached the genome-wide significance threshold only for TD, likely due to the higher heritability of the trait. This SNP explained a moderate fraction (17%) of genetic variance for TD suggesting that the majority of the genetic variation is probably explained by oligogenic effects. In such cases genomic selection may be more efficient than marker-assisted selection for this QTL only.
The favorable homozygote and the heterozygote had lower mortality compared to the unfavorable homozygote for both survival and TD, possibly indicating dominance of the favorable allele. However this is uncertain as the frequency of favorable allele was low (16%). Thus, there is a large room for improvement of natural resistance in rainbow trout populations. Based on the present work, relatively good protection could be achieved by avoiding unfavorable homozygotes in commercial production (for example by restricting sires to be homozygous for the favorable allele).
To learn more about the underlying mechanisms of resistance, we identified the positional candidate genes within a 1000 Kb window of the two most significant SNPs on chromosome 16. The most significant SNP (AX-89969631) that explained the greatest proportion of genetic variance was located in the second intron of an uncharacterized non-coding RNA gene (LOC118939552). The second most significant SNP (AX-89973999) was located in the first intron of the gene encoding PRA1 family protein 3 (Gene ID 110492162). This gene has previously been suggested to be linked to A. salmonicida resistance in turbot36 and a its paralogue gene was listed among positional candidate genes associated to Flavobacterium columnare resistance in rainbow trout33. The PRA1 family protein 3 in humans reduces viral anti-apoptotic activity and thus has an important role in host-mediated apoptosis and immune response37. The region close to SNP AX-89973999 contains another gene identified in the QTL region with the largest effect on resistance to F. columnare33. This gene encodes for a FERM domain-containing protein 4B known as a scaffolding protein regulating epithelial cell polarization38. The gene located between the two most significant SNPs encodes for the NEDD8-activating enzyme E1 catalytic subunit. Neddylation is a post-translational protein modification that effects gene regulation, cell survival, organ development, stress- and immune response39. The blocking of neddylation by the NEDD8 activating enzyme E1 inhibitor has shown to suppress antiviral response in zebrafish and increased the sensitivity to spring viremia of carp virus (SVCV) infection40. In close proximity to the SNP location, we found genes encoding chemokine-like protein TAFA-1 and TAFA-4, both belonging to a family of small secreted proteins. These proteins, distantly related to CC chemokines, are primarily expressed in the central nervous system (CNS) and their biological functions might involve functioning as brain-specific chemokines regulating immune and nervous cells41.
Other genes adjacent to the two most significant SNPs comprise the gene encoding leiomodin-3 and microphthalmia-associated transcription factor (MITF). The leiomodin-3 protein has a role in organization of actin filaments in skeletal muscle in zebrafish42 and MITF regulates the expression of several other genes and thereby the lineage-specific regulation of several cell types including melanocytes, mast cells, and osteoclasts43. The latter gene also plays a role for cell homeostasis, apoptosis and cell cycle44.
More distantly located genes that have been reported to be involved in immune response in different fish species include the forkhead box protein P1-B (FOX B-1)—a member of a large family of transcription factors well-characterized in mammals. FOX B-1 has a pivotal role in human and mouse B-cell development and is involved in the expression of recombination-activating genes (RAG) in pro-B cells and in V(D)J recombination45. In zebrafish, FOX B-1 has shown to be an important transcription factor in the development of CNS and other organs46. In rainbow trout, FOX P1 was found up-regulated after A. salmonicida challenge in the gills, contributing in the regulation of brachial immunity in this species47.
It is likely that several genes have a role in the resistance to furunculosis, some related to immune defense and others engaged in different stages of pathogen-host interaction. It was recently suggested that the epithelial cadherin (cdh1) gene, encoding a protein that bind IPNV virions during virus internalization, is a major determinant of the resistance to IPNV in Atlantic salmon48. This frames that the identification of genes underlying the QTL in disease resistance requires knowledge of the intricate and complex interactions between the host and the pathogen49.
None of the positional candidate genes identified in the present study were targeted in the qPCR analysis. However, the gene expression profile gave an overview of the innate and adaptive immune genes generally involved in bacterial infection. It cannot be excluded that one or more of the positional candidate genes mentioned above play a role in the antibacterial response. We examined the immune gene expression response in three different organs: the gills, as the primary site of bacterial entry; the liver, functioning as the main producer of complement- and acute phase proteins; and lastly the spleen, responsible for antigen presentation and immune regulation50.
The highest transcriptional changes post-exposure took place in the liver and spleen. The gene regulation response in various organs could be related to the level of bacterial transcripts in these organs at 3 dpc. In the gills, where the gene regulation was lower compared to other organs, only a single fish was detected with bacterial gene transcripts. Both liver and spleen, showing a strong gene-expression response, were generally positive for bacterial gene transcripts. It was also evident that fish exhibiting clinical signs reacted much stronger when compared to fish without any visible disease signs. Thus, the magnitude of invasion reflects the strength of the response. The surviving fish had no bacterial transcripts in the gills, liver and spleen, suggesting that these fish are able to clear the infection in these organs. However, further studies should investigate if the survivors were free from infection during the entire challenge period. Re-challenge of survivors would also confirm the pathogen-free status. A. salmonicida has been found in the gut and brain of asymptomatic carrier fish51 suggesting that a low number of pathogens are kept at distance by the innate (SAA, C3, cathelicidins, lysozyme) and adaptive (IgM) immune genes activated in the resistant surviving fish.
The early infection (3 dpc) triggered a strong transcriptional up-regulation of many immune genes in different organs including those encoding pro-inflammatory cytokines IL-1β, IL-6 and TNFα and other cytokines involved in the development of adaptive immunity (IL-8, IL-10, IL-22, IL-17C2, INFγ). The clear up-regulation of previously mentioned immune markers is consistent with earlier studies in trout exposed to A. salmonicida47,52,53 and other bacterial pathogens12,35. Along with the inflammation induced we saw a marked up-regulation of SAA, C3 and several other components of the innate immune system (lysozyme and cathelicidins). IgM was the only immunoglobulin found to be up-regulated from the early state of infection. This confirms previous suggestions that antibodies are key players in protection against A. salmonicida7. We clearly observed a systemic immune response to A. salmonicida infection in trout but it is noteworthy that susceptible fish achieving a high pathogen load and with external clinical signs generally responded stronger than fish resisting the challenge. However, the surviving fish could have resisted the infection based on one or several factors including possible candidate genes associated with specific SNPs, innate immune components and IgM.
Conclusion
This is to our knowledge the first study that aims to identify the genomic regions associated with Aeromonas salmonicida resistance in rainbow trout by using the 57 K SNP array. We found that resistance to A. salmonicida is moderately heritable and showed an oligogenic architecture. We discovered a significant QTL on cromosome 16 and the most significant SNP explained 17% of the genetic variance in resistance to furunculosis. Rainbow trout infected with A. salmonicida exhibited a notable up-regulation of cytokine and innate immune factor genes 3 days after bacterial challenge in gills, liver and spleen, indicating an induction of systematic immune response to the infection. Future GWAS analysis should further validate the oligogenic nature of the resistance to A. salmonicida. In addition, comparative transcriptome analyses could provide more detailed information on gene expression differences between susceptible versus resistant fish and the possible connection with detected candidate genes. Additional gene editing techniques may be applied to validate the functional role of different genes in resistance in trout and other host species.
Materials and methods
Ethics statement
Infection procedures were performed under the license no. 2019-15-0201-01614 issued by the Experimental Animal Inspectorate, Committee for Experimental Animals, Ministry of Environment and Food, Denmark. ARRIVE guidelines and ethical guidelines of the University of Copenhagen were followed securing that fish were monitored every second hour around-the-clock. Fish showing clinical signs (loss of equilibrium, irregular swimming, skin hemorrhages, severe discoloration) were immediately euthanized with overdose (300 mg/l) of tricaine methane sulphonate (MS222, Sigma-Aldrich, Søborg, Denmark) and recorded as mortalities.
Fish
An outbred population of 60 half-sibling families (12–13 fish from each family) of rainbow trout was used for the experiments (Hallesø trout farm, Aquasearch ova ApS, Jutland, Denmark). Disinfected eyed rainbow trout eggs, were transported to Aqua Baltic pathogen-free hatchery54 (Nexø, Denmark) and hatched at 7 °C during 14 days. Fish were reared in 700 l tanks with recirculating municipal water (12 °C) and fed 1% biomass of dry pelleted feed (INICIO 917 BioMar A/S, Brande, Denmark) daily. The juvenile fish (1800 degree-days post-hatch, average body weight 8 g and length 8.5 cm) were transported to the experimental fish facility at the University of Copenhagen (Frederiksberg, Denmark). The fish for the challenge study were allocated in twelve 150 l tanks equipped with internal biofilters (20 l/min EHEIM, Deizisau, Germany), accommodating approximately 70 fish per tank. Fish were acclimatized at 19 °C for 14 days prior the challenge in order to conduct the infection study at temperature similar to outbreak conditions. Fish were fed 1% biomass daily (INICIO 917, BioMar) and the rearing conditions were kept constant at pH 7.6, nitrite < 0.01 mg/l, nitrate < 50 mg/l (Tetra GmbH, Melle, Germany), ammonia < 0.5 mg/l (Hach, Loveland, CO, USA) and 30% water was replenished every day. Non-exposed control fish were kept under corresponding conditions in a separate room during the study.
Challenge
Head kidney swabs from 5 freshly euthanized fish were plated onto 5% blood agar plates (SSI Diagnostica, Hillerød, Denmark) before challenge to confirm the fish were free from bacterial infection55. Fish (n = 798) were exposed to A. salmonicida subsp. salmonicida strain 111129-1/2 by bath challenge. The 48 h bacterial culture (total volume 5 l, 4.7 × 108 cfu/ml) was added (400 ml to each fish tank carrying 30 l aerated water) whereby the fish were exposed to a bacterial concentration of 6.26 × 106 cfu/ml. Fish were kept exposed for 6 h whereafter tanks were filled with tap water to reach the volume of 150 l. Fish were then observed for occurrence of any disease signs for 13 days.
Sampling
Mortality recordings
Mortality was recorded from the exposure time point to 13 days post challenge (dpc). The remaining fish were sampled at 13 dpc and recorded as survivors.
Sampling for genotyping
Samples for genotyping were taken from a total of 798 fish throughout the course of infection. Fish showing clinical signs (653) were sampled as susceptible and at 13 dpc all fish surviving the challenge (145) were sampled as resistant. For DNA-typing, a circular tissue piece (Ø 2.75 mm) was taken from the tail fin of each fish using punching scissors (AgnTho’s AB, Lidingö, Sweden) and transferred to lysis buffer (Vaxxinova Norge, Bergen, Norway) for subsequent DNA purification and genotyping according to Karami et al.12.
Genotyping
The high-density 57 K single nucleotide polymorphism (SNP) chip array developed for rainbow trout28 by ®Affymetrix, San Diego, CA, USA, was used for genotyping. All analysis was conducted according to the Axiom platform Assay-Automated-Workflow-User-Guide56.
Sampling for gene expression analysis
Gills, liver and spleen were sampled from 15 non-exposed control fish at day zero. At 3 dpc samples were obtained from 15 non-exposed control fish, 15 exposed fish showing clinical signs (CS) and 15 exposed fish showing no clinical signs (NCS). At 13 dpc when no fish showed clinical signs of disease, 15 non-exposed control fish and 15 exposed but surviving fish were sampled. Samples were fixed in RNAlater (R0901, Sigma-Aldrich), placed at 4 °C for 24 h and subsequently stored at -20 °C until further processing.
Quantitative RT-PCR (qPCR)
The genes investigated by qPCR analysis were encoding immune-relevant molecules and included SAA, C3, lysozyme, cathelicidins, IFNγ, IgM, IgT, IgD, IL-1β, IL-2, IL-4/13a, IL-8, IL-10, IL-12, IL-17A/F2, IL-17C1, IL-17C2, IL-22, TCRβ, TGFβ, TNFα. Primers and Taq-Man probes applied are listed in Supplementary Table S2 and the gene expression analysis was performed as previously described35. In brief, samples from gills, liver and spleen were homogenized (Tissue-lyser II, Qiagen, Vedbæk, Denmark) and RNA was extracted by GenEluteTM mammalian RNA kit (RTN350, Sigma-Aldrich). The cDNA was synthesized in T100 thermocycler (Biorad, Copenhagen, Denmark) using Oligo d(T)16 primers and TaqMan® reverse transcription reagents (N8080234, Thermo Fischer Scientific, Roskilde, Denmark). Quantitative PCR assays were performed using AriaMx Real-Time PCR machine (G8830A-04R-010, AH diagnostics AS, Denmark). Primers and Taq-Man probes targeting immune-relevant rainbow trout genes were synthesized at TAG Copenhagen AS, Copenhagen, Denmark. The 12.5 µl total volume reactions consisted of 2.5 μl cDNA, 6.25 μl Brilliant III Ultra-Fast QPCR Master Mix (600,881, AH Diagnostics AS, Tilst, Denmark), 1.0 μl primer–probe mixture (10 μM forward primer and reverse primer, 5 μM Taq-Man probe) and 2.75 μl RNase-free water (Invitrogen, Denmark). Reverse transcriptase minus and negative controls were used for every plate setup. The A. salmonicida infection level was monitored by quantifying the bacterial load in different organs using qPCR primers and probes targeting the aopO gene (DQ386862) of A. salmonicida57. The bacterial transcript level was estimated as 107 × 2−∆Cq based on bacterial cDNA as previously described for Yersinia ruckeri35. It should be noted that dead or inactive bacteria is not detected by this method.
Data analysis
Survival
GraphPad Prism version 9.0.0. (GraphPad Software www.graphpad.com) was used to estimate the cumulative mortality rates by Kaplan-Mayer survival analysis.
Gene expression
All qPCR assays exhibited efficiencies within 100% ± 5% and the simplified 2-ΔΔCq method was used for data analysis58. The average of three genes (arp, elf1α and β-actin) was chosen for normalization and as internal calibrator, using NormFinder59. For gene expression analysis at 3 dpc, 2 challenged groups and one control group were compared using one-way ANOVA with Tukey’s test and for analysis at 13 dpc, survivors and control fish were compared with student’s t-test. Minimum twofold regulations were considered substantial and differences between groups were tested with 2-tailed t-test (p < 0.05). Qualitative assessment was applied for the groups that had less than 3 positive Cq values. For that purpose, presence or absence of Cq values was analyzed with nonparametric Mann–Whitney test (p < 0.05).
Genetic analysis
Only fish of high genotype quality were included in the genetic analysis. Therefore, 46 fish were discarded from the study before genetic analysis, resulting in a dataset of 752 fish (out of 798 fish)—138 survivors and 614 dead individuals. The resistance traits investigated were survival and time to death (TD—survival interval between infection and death). SNPs included in the study were restricted to 32 205 loci and showed all three possible genotype clusters (PolyHighRes) by Thermo Fisher Array Power Tools software. Statistical analysis was performed according to our latest QTL study12. Shortly, the genomic relationship matrix (GRM) was computed using the Genome-Wide Complex Trait Analysis (GCTA) software60. The data set was initially analyzed with a simple genomic animal model using restricted maximum likelihood (REML):
Where y is a vector of phenotypes (0/1 for survival or TD), b is the vector of fixed effects (12 different tanks) with associated incidence matrix X, is a vector of additive genetic (polygenic) effects, is the genomic relationship matrix, computed from all high-quality markers, is the polygenic variance, is a vector of random residual effects and is the residual variance. The genetic variance was calculated (assuming that the estimated SNP effect is the actual effect) using a following formula: , where freq is the allele frequency and b is the estimated SNP effect.
Secondly, a leave-one-chromosome-out genome-wide association study (LOCO-GWAS) was performed using the same model as above but extended with individual SNP effects. Here, polygenic effects on all other chromosomes, except the one being currently tested, was accounted for. The rationale using the LOCO-GWAS model is that the model takes into account the potential stratification of the tested population, which is especially relevant to consider in farmed fish with strong relationship structures. In the current model, an animal genetic (polygenic) effect was therefore included, with animal effects having a covariance structure throughout the GRM along with fixed effects on the SNP that is currently tested (all SNPs are tested as separate effects, one-by-one). Thus, the SNP effects were solutions from the linear mixed LOCO-GWAS model which included a fixed overall mean, a fixed regression using the SNP being tested as a covariate (the estimated SNP effect is the solution for this regression coefficient) and a random polygenic animal effect (with GRM estimated from all chromosomes except the one being tested).
Using a linear model, heritability was estimated on the observed scale. The heritability estimates for survival were transformed to the underlying liability scale using a build-in option of GCTA software based on the theory and formulas presented in Lee et al.61.
For the SNPs to be significantly associated with resistance, Bonferroni correction was applied (i.e. , where N refers to the number of SNPs).
Supplementary Information
Acknowledgements
The authors are indebted to the staff members of Aqua Baltic Salmon Hatchery in Nexø, Bornholm, Denmark, for hatching and rearing the fish for the present study. We would also like to acknowledge the laboratory technician Kári K. Mouritsen at the National Institute of Aquatic Resources for cultivating the bacterial broth. This study was funded by the Innovation Fund Denmark IFD grant no. 8090-00002B.
Author contributions
K.B. and T.N. designed the experimental work. M.H.M, A.M.K, J.Ø., S.Z., R.M.J., H.M., L.V.G.J. performed the experiment. I.D. provided the pathogen. J.Ø. and P.W.K. analyzed the data. M.H.M. wrote the manuscript. All authors provided feedback and edited the manuscript. All authors have read and approved the final manuscript.
Data availability
The datasets generated and data analysed during the current study are made available from the corresponding author on request.
Competing interests
Jørgen Ødegård and Torben Nielsen were employed by companies AquaGen and Aquasearch ova ApS, respectively. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-97437-7.
References
- 1.Menanteau-Ledouble S, Kumar G, Saleh M, El-Matbouli M. Aeromonas salmonicida: Updates on an old acquaintance. Dis. Aquat. Organ. 2016;120:49–68. doi: 10.3354/dao03006. [DOI] [PubMed] [Google Scholar]
- 2.Austin B, Austin D. Aeromonadaceae representative (Aeromonas salmonicida) In: Austin B, editor. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish. Springer; 2012. pp. 147–228. [Google Scholar]
- 3.Derome NJ, Gauthier BS, Llewellyn M. Bacterial opportunistic pathogens in fish. In: Hurst C, editor. The Rasputin Effect: When Commensals and Symbionts Become Parasitic. Advances in Environmental Microbiology. Springer; 2016. pp. 81–108. [Google Scholar]
- 4.Tam B, Gough WA, Tsuji L. The impact of warming on the appearance of furunculosis in fish of the James Bay region, Quebec, Canada. Region. Environ. Change. 2011;11:123–132. doi: 10.1007/s10113-010-0122-8. [DOI] [Google Scholar]
- 5.De Silva SS, Soto D. Climate Change and Aquaculture: Potential Impacts, Adaptation and Mitigation. Climate Change Implications for Fisheries and Aquaculture: Overview of Current Scientific Knowledge. FAO Fisheries and Aquaculture Technical Paper (530); 2009. pp. 151–212. [Google Scholar]
- 6.Midtlyng PJ, Reitan LJ, Speilberg L. Experimental studies on the efficacy and side-effects of intraperitoneal vaccination of Atlantic salmon (Salmo salar L.) against furunculosis. Fish Shellfish Immunol. 1996;6:335–350. doi: 10.1006/fsim.1996.0034. [DOI] [Google Scholar]
- 7.Marana MH, et al. Positive correlation between Aeromonas salmonicida vaccine antigen concentration and protection in vaccinated rainbow trout Oncorhynchus mykiss evaluated by a tail fin infection model. J. Fish Dis. 2017;40:507–516. doi: 10.1111/jfd.12527. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen K, et al. Surveillance of health status on eight marine rainbow trout, Oncorhynchus mykiss (Walbaum), farms in Denmark in 2006. J. Fish Dis. 2008;31:659–667. doi: 10.1111/j.1365-2761.2008.00941.x. [DOI] [PubMed] [Google Scholar]
- 9.Ødegård J, Baranski M, Gjerde B, Gjedrem T. Methodology for genetic evaluation of disease resistance in aquaculture species: challenges and future prospects. Aquac. Res. 2011;42:103–114. doi: 10.1111/j.1365-2109.2010.02669.x. [DOI] [Google Scholar]
- 10.Houston RD, et al. major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar) Genetics. 2008;178:1109–1115. doi: 10.1534/genetics.107.082974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moen T, Baranski M, Sonesson AK, Kjøglum S. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics. 2009;10:368. doi: 10.1186/1471-2164-10-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karami AM, et al. A major QTL for resistance to Vibrio anguillarum in rainbow trout. Front. Genet. 2020 doi: 10.3389/fgene.2020.607558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vallejo RL, et al. Detection of QTL in rainbow trout affecting survival when challenged with Flavobacterium psychrophilum. Mar. Biotechnol. 2014;16:349–360. doi: 10.1007/s10126-013-9553-9. [DOI] [PubMed] [Google Scholar]
- 14.Boison SA, Gjerde B, Hillestad B, Makvandi-Nejad S, Moghadam HK. Genomic and transcriptomic analysis of amoebic gill disease resistance in Atlantic Salmon (Salmo salar L.) Front. Genet. 2019;10:68. doi: 10.3389/fgene.2019.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaafar R, et al. Quantitative trait loci (QTL) associated with resistance of rainbow trout Oncorhynchus mykiss against the parasitic ciliate Ichthyophthirius multifiliis. J. Fish Dis. 2020;43:1591–1602. doi: 10.1111/jfd.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Embody GC, Hayford CO. The advantage of rearing brook trout fingerling from selected breeders. Trans. Am. Fish. Soc. 1925;55:135–142. doi: 10.1577/1548-8659(1925)55[135:TAORBT]2.0.CO;2. [DOI] [Google Scholar]
- 17.Wolf LE. Development of disease-resistant strains of fish. Trans. Am. Fish. Soc. 1954;83:342–349. doi: 10.1577/1548-8659(1953)83[342:DODSOF]2.0.CO;2. [DOI] [Google Scholar]
- 18.Snieszko SF, Dunbar CE, Bullock GL. Resistance to ulcer disease and furunculosis in Eastern brook trout, Salvelinus fontinalis. Prog. Fish Cult. 1959;21:111–116. doi: 10.1577/1548-8659(1959)21[111:RTUDAF]2.0.CO;2. [DOI] [Google Scholar]
- 19.Ehlinger NF. Selective breeding of trout for resistance to furunculosis. NY Fish Game J. 1977;24:25–36. [Google Scholar]
- 20.Cipriano RC, Marchant D, Jones TE, Schachte JH. Practical application of disease resistance: A brook trout fishery selected for resistance to furunculosis. Aquaculture. 2002;206:1–17. doi: 10.1016/S0044-8486(01)00863-8. [DOI] [Google Scholar]
- 21.Gjedrem T, Gjøen HM. Genetic variation in susceptibility of Atlantic salmon, Salmo salar L., to furunculosis, BKD and cold water vibriosis. Aquac. Res. 1995;26:129–134. doi: 10.1111/j.1365-2109.1995.tb00892.x. [DOI] [Google Scholar]
- 22.Gjøen HM, Refstie T, Ulla O, Gjerde B. Genetic correlations between survival of Atlantic salmon in challenge and field tests. Aquaculture. 1997;158:277–288. doi: 10.1016/S0044-8486(97)00203-2. [DOI] [Google Scholar]
- 23.Langefors A, Lohm J, Grahn M, Andersen O, von Schantz T. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. Biol. Sci. 2001;268:479–485. doi: 10.1098/rspb.2000.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimholt U, et al. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Niu C, Storset A, Bøgwald J, Dalmo RA. Comparison of Aeromonas salmonicida resistant and susceptible salmon families: A high immune response is beneficial for the survival against Aeromonas salmonicida challenge. Fish Shellfish Immunol. 2011;31:1–9. doi: 10.1016/j.fsi.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Cipriano RC. Resistance of salmonids to Aeromonas salmonicida: relation between agglutinins and neutralizing activities. Trans. Am. Fish. Soc. 1983;112:95–99. doi: 10.1577/1548-8659(1983)112<95:ROSTAS>2.0.CO;2. [DOI] [Google Scholar]
- 27.Hollebecq M-G, Faivre B, Bourmaud C, Michel C. Spontaneous bactericidal and complement activities in serum of rainbow trout (Oncorhynchus mykiss) genetically selected for resistance or susceptibility to furunculosis. Fish Shellfish Immunol. 1995;5:407–426. doi: 10.1006/fsim.1995.0040. [DOI] [Google Scholar]
- 28.Palti Y, et al. The development and characterization of a 57K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 2015;15:662–672. doi: 10.1111/1755-0998.12337. [DOI] [PubMed] [Google Scholar]
- 29.Vallejo RL, et al. Genomic selection models double the accuracy of predicted breeding values for bacterial cold water disease resistance compared to a traditional pedigree-based model in rainbow trout aquaculture. Genet. Sel. Evol. 2017;49:17. doi: 10.1186/s12711-017-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallejo RL, et al. Similar genetic architecture with shared and unique quantitative trait loci for bacterial cold water disease resistance in two rainbow trout breeding populations. Front. Genet. 2017 doi: 10.3389/fgene.2017.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, et al. Retrospective evaluation of marker-assisted selection for resistance to bacterial cold water disease in three generations of a commercial rainbow trout breeding population. Front. Gene. 2018 doi: 10.3389/fgene.2018.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez FH, et al. Genome-wide association analysis for resistance to infectious pancreatic necrosis virus identifies candidate genes involved in viral replication and immune response in rainbow trout (Oncorhynchus mykiss) G3. 2019;9:2897–2904. doi: 10.1534/g3.119.400463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva RMO, et al. Whole-genome mapping of quantitative trait loci and accuracy of genomic predictions for resistance to columnaris disease in two rainbow trout breeding populations. Genet. Sel. Evol. 2019;51:42. doi: 10.1186/s12711-019-0484-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallejo RL, et al. Genome-wide association analysis and accuracy of genome-enabled breeding value predictions for resistance to infectious hematopoietic necrosis virus in a commercial rainbow trout breeding population. Genet. Sel. Evol. 2019;51:47. doi: 10.1186/s12711-019-0489-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo S, et al. Immune gene expression and genome-wide association analysis in rainbow trout with different resistance to Yersinia ruckeri infection. Fish Shellfish Immunol. 2020;106:441–450. doi: 10.1016/j.fsi.2020.07.023. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Ramilo ST, et al. QTL detection for Aeromonas salmonicida resistance related traits in turbot (Scophthalmus maximus) BMC Genomics. 2011;12:541. doi: 10.1186/1471-2164-12-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li LY, Shih HM, Liu MY, Chen JY. The cellular protein PRA1 modulates the anti-apoptotic activity of Epstein-Barr virus BHRF1, a homologue of Bcl-2, through direct interaction. J. Biol. Chem. 2001;276:27354–27362. doi: 10.1074/jbc.M103821200. [DOI] [PubMed] [Google Scholar]
- 38.Klarlund JK, et al. Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. J. Biol. Chem. 2001;276:40065–40070. doi: 10.1074/jbc.M105260200. [DOI] [PubMed] [Google Scholar]
- 39.Brockmann MM, et al. Neddylation regulates excitatory synaptic transmission and plasticity. Sci. Rep. 2019;9:17935. doi: 10.1038/s41598-019-54182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G, et al. Neddylation facilitates the antiviral response in zebrafish. Front. Immunol. 2019 doi: 10.3389/fimmu.2019.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tom Tang Y, et al. TAFA: A novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics. 2004;83:727–734. doi: 10.1016/j.ygeno.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Yuen M, et al. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J Clin. Invest. 2014;124:4693–4708. doi: 10.1172/JCI75199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martina JA, Diab HI, Li H, Puertollano R. Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell Mol. Life Sci. 2014;71:2483–2497. doi: 10.1007/s00018-014-1565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang T, et al. Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1. J. Cell Sci. 2015;128:2938–2950. doi: 10.1242/jcs.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu H, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat. Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 46.Cheng L, et al. Molecular cloning, characterization, and developmental expression of foxp1 in zebrafish. Dev. Genes Evol. 2007;217:699–707. doi: 10.1007/s00427-007-0177-9. [DOI] [PubMed] [Google Scholar]
- 47.Rebl A, et al. Transcriptome profiling reveals insight into distinct immune responses to Aeromonas salmonicida in gill of two rainbow trout strains. Mar. Biotechnol. 2014;16:333–348. doi: 10.1007/s10126-013-9552-x. [DOI] [PubMed] [Google Scholar]
- 48.Moen T, et al. Epithelial cadherin determines resistance to infectious pancreatic necrosis virus in Atlantic Salmon. Genetics. 2015;200:1313–1326. doi: 10.1534/genetics.115.175406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraslin C, et al. Combining multiple approaches and models to dissect the genetic architecture of resistance to infections in fish. Front. Genet. 2020 doi: 10.3389/fgene.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Secombes CJ, Wang T. The innate and adaptive immune system of fish. In: Austin B, editor. Infectious Disease in Aquaculture: Prevention and Control. Woodhead Publishing Limited; 2012. pp. 3–68. [Google Scholar]
- 51.Bartkova S, Kokotovic B, Skall HF, Lorenzen N, Dalsgaard I. Detection and quantification of Aeromonas salmonicida in fish tissue by real-time PCR. J. Fish Dis. 2017;40:231–242. doi: 10.1111/jfd.12505. [DOI] [PubMed] [Google Scholar]
- 52.Brietzke A, et al. Aeromonas salmonicida infection only moderately regulates expression of factors contributing to toll-like receptor signaling but massively activates the cellular and humoral branches of innate immunity in rainbow trout (Oncorhynchus mykiss) J. Immunol. Res. 2015;2015:901015. doi: 10.1155/2015/901015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Causey DR, et al. High-throughput proteomic profiling of the fish liver following bacterial infection. BMC Genom. 2018;19:719–719. doi: 10.1186/s12864-018-5092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xueqin J, Kania PW, Buchmann K. Comparative effects of four feed types on white spot disease susceptibility and skin immune parameters in rainbow trout, Oncorhynchus mykiss (Walbaum) J. Fish Dis. 2012;35:127–135. doi: 10.1111/j.1365-2761.2011.01329.x. [DOI] [PubMed] [Google Scholar]
- 55.Dalsgaard I, Madsen L. Bacterial pathogens in rainbow trout, Oncorhynchus mykiss (Walbaum), reared at Danish freshwater farms. J. Fish Dis. 2000;23:199–209. doi: 10.1046/j.1365-2761.2000.00242.x. [DOI] [Google Scholar]
- 56.Axiom™. Assay 96-Array Format Automated Workflow. User Quide. https://assets.thermofisher.com/TFS-Assets/LSG/manuals/702963_Final_Ax_96FX_UG.pdf 2.0).
- 57.Fernández-Álvarez C, González SF, Santos Y. Development of a SYBR green I real-time PCR assay for specific identification of the fish pathogen Aeromonas salmonicida subspecies salmonicida. Appl. Microbiol. Biotechnol. 2016;100:10585–10595. doi: 10.1007/s00253-016-7929-2. [DOI] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.Can-04-0496. [DOI] [PubMed] [Google Scholar]
- 60.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and data analysed during the current study are made available from the corresponding author on request.