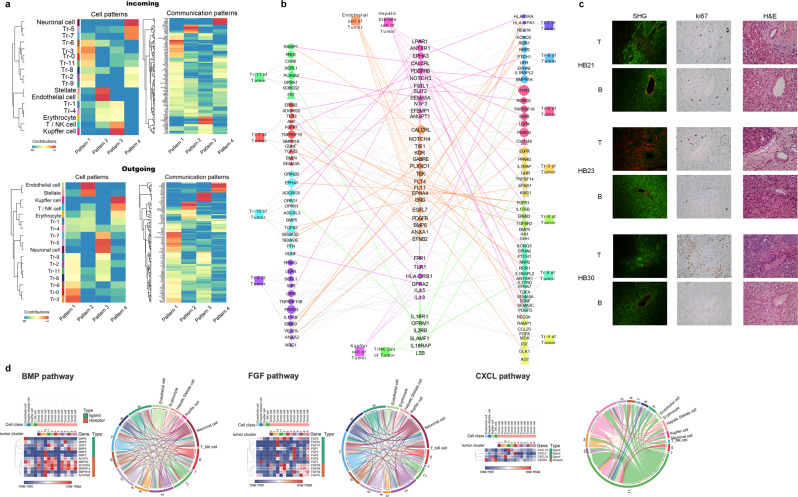
Fig. 7. Differential signaling between tumor cell clusters and microenvironment cells.
a Signaling pathways were predicted using CellChat to show interaction patterns between tumor subclusters and microenvironment cells. Four main patterns of ingoing and outgoing signals were identified, and associated pathways shown. b A network of signaling interactions of upregulated genes between tumor cell clusters and supporting tumor microenvironment cells shows crosstalk between cells in tumor. Gene–gene interactions were inferred using ToppCluster and interaction pairs between tumor cells and microenvironment cells were highlighted. Node size represents normalized expression levels of genes in each cell type. Node shape represents ligands (triangle) and receptors (circle). c Second harmonic generation (SHG) microscopy of tumor and background liver. Left to right, SHG (red—SHG, green—autofluorescence), KI67, and H&E staining demonstrates disorganized vessel structure in tumor samples. 20X Plan APO, 1024 × 1024 resolution, scale bar 50 µM. d Heatmaps and CellChat chord diagrams of upregulated pathways with indicated receptor/ligand interactions are highlighted. Extensive BMP, FGF, and CXCL signaling networks are shown. Lowly expressed genes in each pathway (maximal row values less than 0.5) were removed from the heatmap.