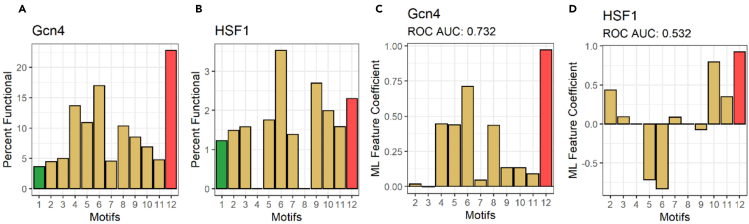
Figure 1.
SLiMs of tADs are localized predominantly in nonfunctional subpools and perform poorly as an ML feature set
(A and B) Y axis: percent of tAD functional sequences in the library. X axis: (1) entire library. Sequences containing SLiMs of (2) p53; (3) RelA/p65; (4) CREBZF; (5) AR; (6) ANACO13, (7) EKLF; (8) Gcn4; (9) 9 aa tAD consensus (stringent); (10) 9 aa tAD consensus (moderate); (11) 9 aa tAD consensus (lenient); (12) all sequences devoid of K, R, and H.
(C and D) Y axis: ML feature coefficients of SLiMs, performed as features using ML ridge regression to predict function and trained on (C) Gcn4 library or (D) HSF1. X axis: SLiM source as in panel (A) and (B).