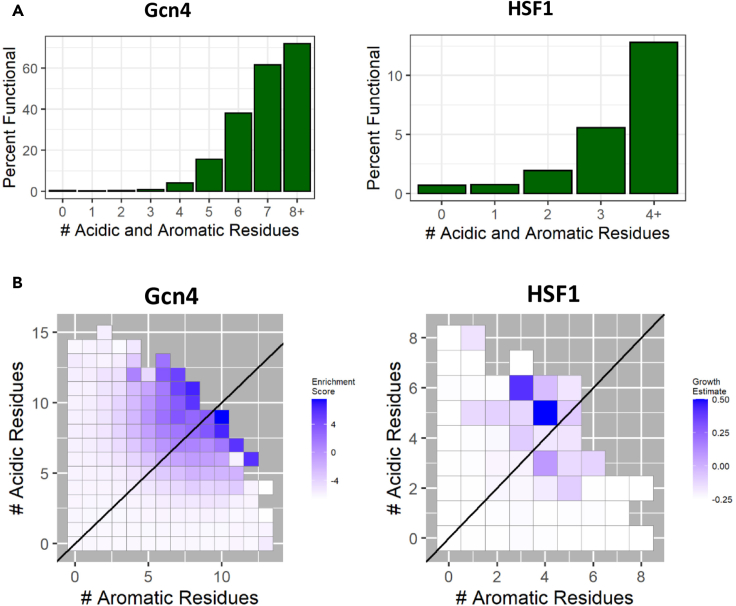
Figure 4.
Multiple aromatic and acidic residues within an individual tAD increase both the probability and the level of function
(A) Y axis: percent of functional sequences in the library. X axis: the number of acidic (D and E) and aromatic (W, F, and Y) amino acids within individual tAD sequences.
(B). Y axis: count of acidic amino acids within an individual tAD sequence. X axis: count of aromatic amino acids within an individual tAD sequence. The enrichment score for both Gcn4 and HSF1 is the average of the scores for all sequences with the specified number of acidic and aromatic amino acids. The diagonal line indicates an equal number of aromatic and acidic amino acids.