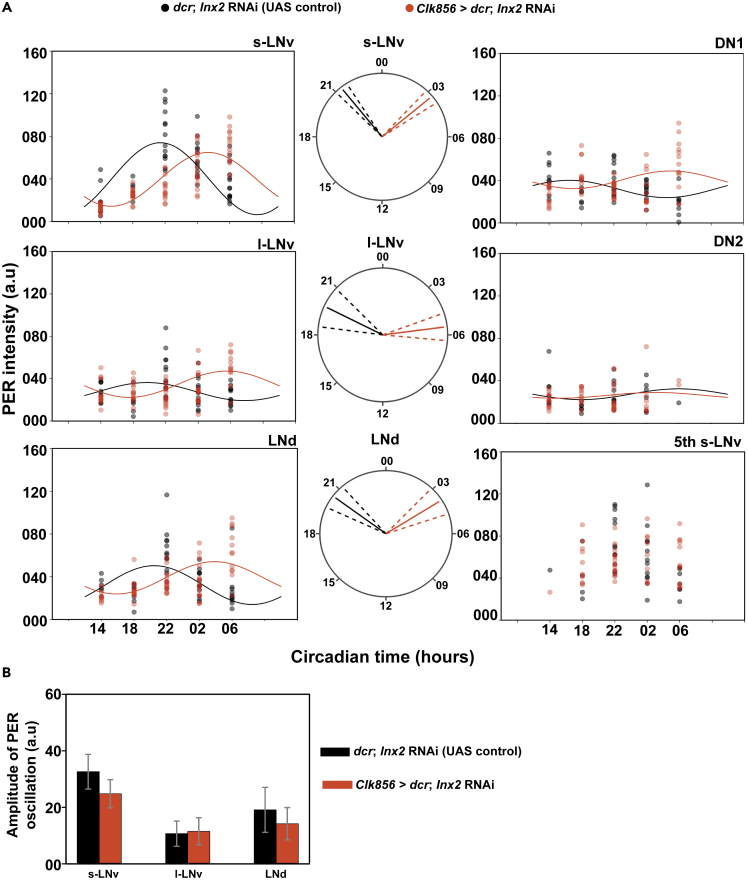
Figure 6.
Knockdown of Innexin2 delays the oscillation of PER in clock neuronal subsets
(A) Scatterplots of PER staining intensities in each of the bilaterally located six distinct neuronal clusters of the circadian pacemaker network of both the control (dcr; Inx2 RNAi) and experimental (Clk856 > dcr; Inx2 RNAi) flies plotted at different time-points over a 24-hr cycle on third day of DD. Each dot represents the mean PER intensity value averaged over both the hemispheres of one brain. The black and red lines are the best fit COSINE curve from the parameters that were extracted using COSINOR analysis. Polar-plots are depicting the acrophase of PER oscillation in control (dcr; Inx2 RNAi) (black lines) and experimental flies (Clk856 > dcr; Inx2 RNAi) (red lines) in 3 distinct neuronal clusters of the circadian pacemaker network where both control and experimental genotypes show a significant 24-hr rhythm in COSINOR analysis. The acrophase values obtained after COSINOR curve fitting are shown as solid lines and the error (95% CI values) is depicted as dashed lines around the mean for all the cell types. Non-overlapping error values indicate that phase values of experimental flies are significantly different from controls as seen in the case of s-LNv, l-LNv, and LNd.
(B) Amplitude values obtained from COSINOR curve fits are plotted for control and experimental flies for those cell groups which show significant 24-hr rhythms (s-LNv, l-LNv and LNd). Error bars are 95% CI values calculated from the standard error obtained from COSINOR analysis. Overlapping error bars indicate that amplitude values of experimental flies are not significantly different from controls. COSINOR analysis was implemented using the CATCosinor function from the CATkit package written for R (Lee Gierke and Cornelissen, 2016). See Table 3; Figures S8 and S9 for more details. n > 7 brain samples both in case of control and experimental flies in most cell types (s-LNv, l-LNv, LNd, DN1) and all time points except for DN2.