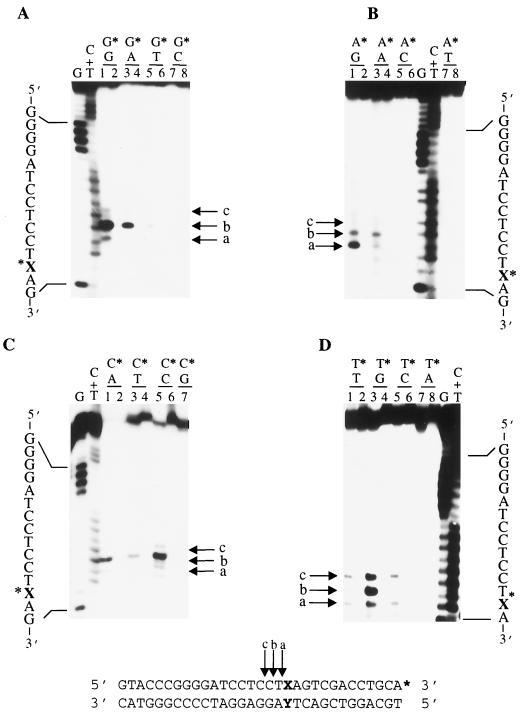
FIG. 1.
GΔ228-Uve1p recognizes 12 different base mismatch combinations. The 3′-end-labeled oligonucleotide series X/Y-31mer (sequence given at bottom; asterisk indicates labeled strand and labeled terminus) was used to assess Uve1p cleavage activity on 16 different base pair and base mispair combinations (Table 1). Base mispairs are indicated above numbered lanes, with asterisks denoting bases on the labeled strand for G series (A), A series (B), C series (C), and T series (D) treated with purified GΔ228-Uve1p (odd-numbered lanes) or mock reactions (even-numbered lanes). Reaction products were analyzed on DNA sequencing-type gels. Arrows indicate Uve1p cleavage sites immediately (arrow a) and one (arrow b) and two (arrow c) nucleotides 5′ to the mismatch site. G and C+T base-specific chemical cleavage DNA sequencing ladders were run in adjacent lanes as nucleotide position markers.