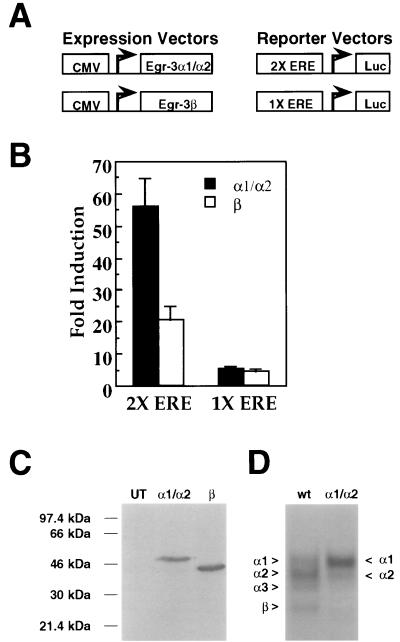
FIG. 5.
Transcriptional activity of Egr3 isoforms. (A) Schematic representation of the expression and reporter vectors used in the reporter assays. The 1X ERE and 2X ERE reporter constructs contained either one or two copies of an ERE (GCG GGG GCG) upstream of the firefly luciferase gene. (B) Bar graph showing fold induction in luciferase activity induced by transfection with either CMVEgr3M49A/M106A (encoding Egr3α1/α2) or CMVEgr3Δ1-105/M129L (encoding Egr3β) over background reporter activity (taken from extracts of hEK-293 cell transfected with an empty CMV vector) for the 2X (left) and 1X (right) EREs. Error bars represent standard errors of the means. Statistical analysis of these data demonstrate that the Egr3 constructs tested differ significantly in their effects on the 2X ERE but not the 1X ERE (P < 0.0005 and P > 0.5, respectively; Student’s t test). (C) Immunoblot of untransfected hEK-293 cells run adjacent to hEK-293 cells transfected with 5 μg of CMVEgr3M49A/M106A (encoding Egr3α1/α2) or CMVEgr3Δ1-105/M129L (encoding Egr3β) probed with αEgr3-INT (1:500 dilution). (D) Representative gel shift experiment using a 32P-labeled ERE oligonucleotide probe on extracts of hEK-293 cells untransfected (UT) or transfected with 5 μg of CMVEgr3wt (wt [wild type]) or CMVEgr3M49A/M106A (α1/α2).