In a recent article by Garg et al., 1 serum levels of five microRNAs (miRs) were monitored in intensive care unit patients with severe COVID‐19 pneumonia requiring invasive ventilation. The miR levels were compared to patients with severe influenza resulting in acute respiratory distress syndrome and healthy controls. For miR‐21, a microRNA that is associated with cardiac fibroblast dysfunction, 1 the authors observed elevated levels in acute COVID‐19 patients and positive associations (P ≲ 0.05) with the number of intensive care unit days on ventilation or vasopressor, dialysis and extracorporeal membrane oxygenation. A negative association was observed with lactate dehydrogenase levels.
miR‐21 is an abundant miR implicated in different regulatory pathways and exhibiting altered levels in circulation in neoplastic and other diseases; insight into its actions might lead to new strategies for therapeutic intervention. 1 , 2 Notably, coronaviruses have binding (target) sites for miR‐21. SARS‐CoV‐2, or inflammatory or other affected processes, might thus alter circulating miR‐21 levels.
An approximately 65 kb region of chromosome 3, in band 3p21.31, is the prime locus stably associated with COVID‐19 severity. 3 It spans a haplotype block, i.e. a segment of DNA that is typically inherited as a whole, showing almost no trace of recombination,
and often exhibits few block variants (haplotypes). The risk haplotype is most frequent (≈30%; Figure 1A ) in South Asian populations and was apparently the major haplotype in Neanderthals. 3 It covers a partly intronic segment of the LZTFL1 gene and DNA upstream of the gene SLC60A2. Some 20 single nucleotide polymorphism (SNP) variants distributed along the haplotype block show maximal effect 3 , 4 and almost identical minor allele frequencies (Figure 1A ), thus constituting a checklist of candidate causal SNPs.
Figure 1.
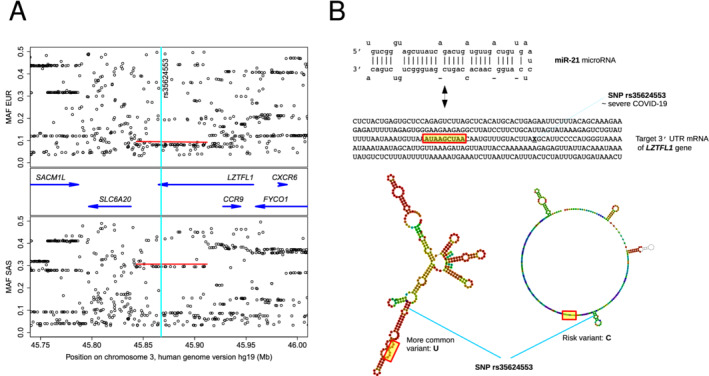
Natural common genetic variation in a COVID‐19 severity‐associated region on chromosome 3 and its potential influence on post‐transcriptional inhibition of LZTFL1 via miR‐21 binding. (A) Genes (blue arrows) and minor allele frequencies (MAF) in two 1000 Genomes Project superpopulations, of European (EUR) and South Asian (SAS) ancestry, showing the risk block's equivalent COVID‐19 associated single nucleotide polymorphisms (SNPs) as rows of points under red bars. Vertical turquoise stripe indicates location of SNP rs35624553, near the border of a functional miR‐21 target site (B), where the variation is predicted to have a sizable effect on structural context (B; RNAfold centroid predictions, with approximate target seed region shown in yellow) and binding efficacy (see main text).
Each possible combination of these SNPs corresponds to a hypothesis of functional contribution(s) to the COVID‐19 symptoms. One of the candidate SNPs, rs17713054, is apparently located in regulatory DNA of SLC60A2 overlapping a CEBP transcription factor binding site and may thus contribute to COVID‐19 severity, since the gene's product SIT1 interacts functionally with the SARS‐CoV‐2 cell surface receptor angiotensin‐converting enzyme 2. 4 The tight coupling (high linkage disequilibrium) of the haplotype block extends into the upstream gene LZTFL1, where it includes other, covarying SNPs such as rs35624553, which could also have an effect on the phenotype (Figure 1B ). SNP rs35624553 is located in transcribed, untranslated 3′ regulatory RNA, less than 20 nt from the seed sequence of a multiply predicted, confirmed miR‐21 target site that has been studied in relation to cancer. 2 A predicted effect of the SNP on the structural landscape embedding the target RNA prior to the encounter with miR‐21 is shown in Figure 1B ; as the SNP is close to the target, differences also in miR‐21 binding efficiency or efficacy are anticipated.
A first working hypothesis for subsequent testing might propose that either the risk allele and/or increased levels of miR‐21 promote down‐regulation of the LZTFL1 gene product, the leucine zipper transcription factor‐like 1 protein, an inhibitor of MAPK signalling. Lowered levels of LZTFL1 would then permit the epithelial–mesenchymal transition (EMT). 2 , 5 EMT has been observed in development, cancers, organ fibrosis, wound healing and inflammation. 2 , 5 Preliminary immunohistochemical and other findings are compatible with a role of EMT or EMT‐like processes also in COVID‐19 or resulting fibrosis (BioRxiv: 2020.11.09.374769 and 2020.05.28.122291v2). Coupling haplotype and cognate microRNA binding studies can identify testable hypotheses to further understand severe COVID‐19 symptoms.
Funding
This work was funded by the Universidad del Rosario, Bogotá via an intramural Big Grant.
References
- 1. Garg A, Seeliger B, Derda AA, Xiao K, Gietz A, Scherf K, Sonnenschein K, Pink I, Hoeper MM, Welte T, Bauersachs J, David S, Bär C, Thum T. Circulating cardiovascular microRNAs in critically ill COVID‐19 patients. Eur J Heart Fail 2021;23:468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang H, Tan Z, Hu H, Liu H, Wu T, Zheng C, Wang X, Luo Z, Wang J, Liu S, Lu Z, Tu J. microRNA‐21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 2019;19:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zeberg H, Pääbo S. The major genetic risk factor for severe COVID‐19 is inherited from Neanderthals. Nature 2020;587:610–612. [DOI] [PubMed] [Google Scholar]
- 4. Wang A, Chiou J, Poirion OB, Buchanan J, Valdez MJ, Verheyden JM, Hou X, Kudtarkar P, Narendra S, Newsome JM, Guo M, Faddah DA, Zhang K, Young RE, Barr J, Sajti E, Misra R, Huyck H, Rogers L, Poole C, Whitsett JA, Pryhuber G, Xu Y, Gaulton KJ, Preissl S, Sun X; NHLBI LungMap Consortium . Single‐cell multiomic profiling of human lungs reveals cell‐type‐specific and age‐dynamic control of SARS‐CoV2 host genes. Elife 2020;9:e62522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wei Q, Chen ZH, Wang L, Zhang T, Duan L, Behrens C, Wistuba II, Minna JD, Gao B, Luo JH, Liu ZP. LZTFL1 suppresses lung tumorigenesis by maintaining differentiation of lung epithelial cells. Oncogene 2016;35:2655–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]


