Figure 1.
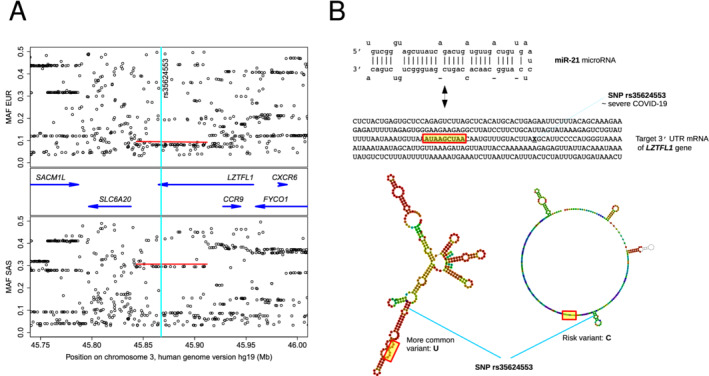
Natural common genetic variation in a COVID‐19 severity‐associated region on chromosome 3 and its potential influence on post‐transcriptional inhibition of LZTFL1 via miR‐21 binding. (A) Genes (blue arrows) and minor allele frequencies (MAF) in two 1000 Genomes Project superpopulations, of European (EUR) and South Asian (SAS) ancestry, showing the risk block's equivalent COVID‐19 associated single nucleotide polymorphisms (SNPs) as rows of points under red bars. Vertical turquoise stripe indicates location of SNP rs35624553, near the border of a functional miR‐21 target site (B), where the variation is predicted to have a sizable effect on structural context (B; RNAfold centroid predictions, with approximate target seed region shown in yellow) and binding efficacy (see main text).
