Fig. 1.
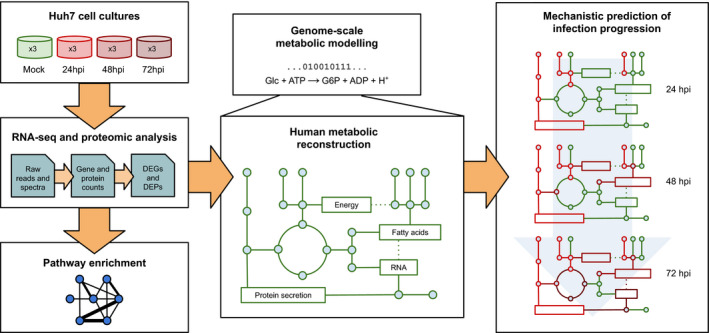
Workflow followed in the study. Time series RNA‐seq and proteomic data from infected Huh7 cells sampled at 24, 48 and 72 hours post‐infection (hpi) were analysed and used to identify DEGs, DEPs and dysregulated pathways compared with uninfected cultures. Results from RNA‐seq and proteomic analysis were used to inform a genome‐scale metabolic reconstruction of human metabolism, which was expanded to include translation and secretion pathways for a range of immune proteins alongside a SARS‐CoV‐2 biomass objective function. Gene and protein expression counts were thus incorporated in the baseline model to capture time‐dependent changes in metabolic activity by estimating DARs with respect to uninfected Huh7 cells.
