Figure 4.
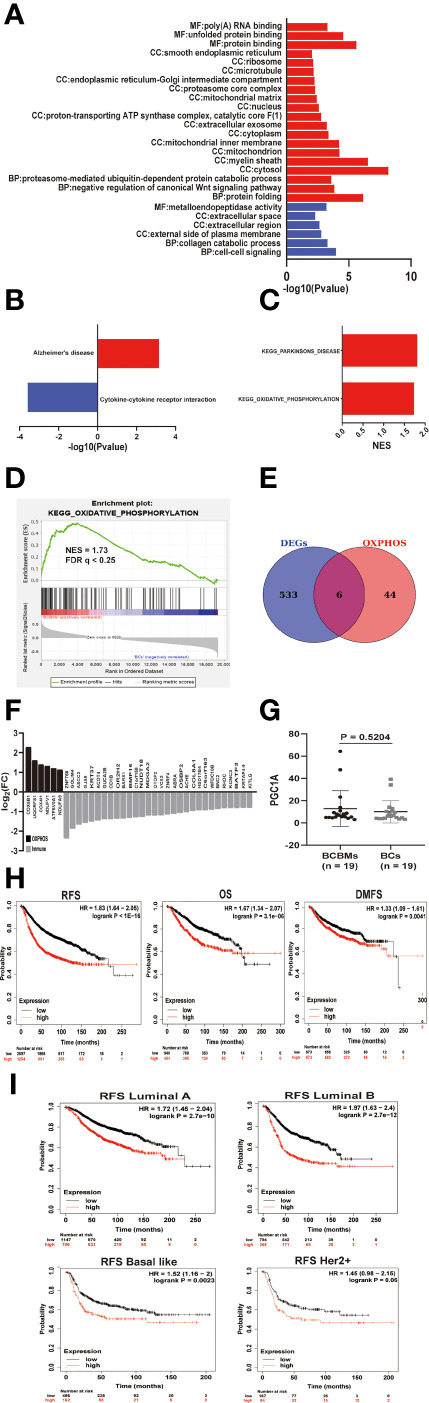
The metabolic features of BCBMs and survival analysis. (A) Gene ontology enrichment analysis of up-regulated (red) and down regulated (blue) genes in BCBMs (n = 19) versus BCs (n = 19) (GSE43837) (P < 0.05; FDR < 0.25). MF: Molecular function, CC: Cell component, BP: Biological process. (B) KEGG enrichment analysis of upregulated and down regulated gene sets in BCBMs (n = 19) versus BCs (n = 19) (GSE43837) (P < 0.05; FDR < 0.25). (C) GSEA analysis demonstrating all KEGG metabolism significantly altered (p < 0.05, FDR < 0.25) in BCBMs (n = 19) versus BCs (n = 19) (GSE43837). The normalized enrichment score (NES) forms the x-axis. Upregulated gene sets are shown in red. No down regulated gene sets met the criteria for statistical significance. (D) GSEA analysis enrichment plot demonstrating significant enrichment of OXPHOS gene set in BCBMs (n = 19) versus BCs (n = 19) (GSE43837). NES and FDR q are listed on the enrichment plot. (E) Venn diagram of DEGs and OXPHOS core enrichment genes obtained by GSEA. A total of 6 overlapping genes were obtained. The OXPHOS core enrichment genes obtained by GSEA are provided in Supplementary Table 6. The 6 overlapping genes are provided in Supplementary Table 7. (F) Bar graph showing log2(FC) values for differentially expressed OXPHOS- and immune-associated genes in BCBMs (n = 19) relative to BCs (n = 19). (G) Comparison of CPM for PGC1A RNA expression between BCBMs (n = 19) and BCs (n = 19) from GSE43837. Lines represent mean ± SD, and each dot represents a single sample. Significance was determined via Wilcoxon rank-sum test. (H) Prognostic significances of OP-metagene in patients with breast cancer were shown based on the KM plotter database. RFS, relapse‐free survival; OS, overall survival; DMFS, distance metastasis free survival; and HR, hazard ratio. The P values were determined using a log-rank test. (I) Prognostic significances of OP-metagene in patients with various breast cancer subtypes were shown based on the KM plotter database.
