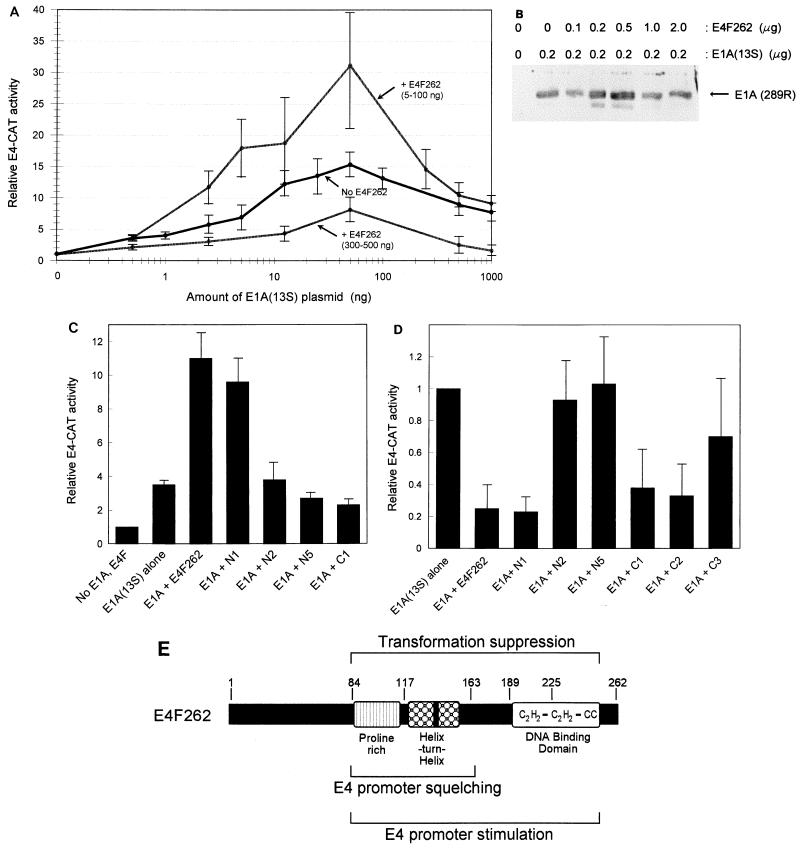
FIG. 4.
Identification of regions in p50E4F that are required for regulation of the E4 promoter. (A) Stimulation and squelching of the E4 promoter by E4F262 and E1A(13S) in NIH 3T3 cells. The relative CAT activity from pE4-CAT1 was determined after cotransfection with increasing amounts of pCMV-E1A(13S) (0.5 to 1,000 ng) and pCMVs-E4F262 (5 to 500 ng). CMV promoter concentrations were kept at a constant level in all transfections by addition of pCMV4. CAT activities normalized for protein concentration, and internal control activities (when included) were divided by the basal CAT activity obtained in the absence of E1A and E4F262 to calculate relative E4-CAT activity. Each individual data point was derived from 3 to 7 independent experiments performed in duplicate; all data points were derived from a total of 12 independent experiments. Standard deviations are shown by thin error bars. Each data point showing p50E4F-mediated stimulation [+ E4F262 (5-100 ng)] or squelching [+ E4F262 (300-500 ng)] was derived from the highest CAT activity level obtained within the specified concentration range of pCMVs-E4F262 for each individual amount of pCMV-E1A(13S). (B) E4F262 does not significantly alter pCMV-E1A(13S) expression. Shown are Western blot of lysates from NIH 3T3 cells cotransfected with 0.2 μg of pCMV-E1A(13S) and the specified amounts of pCMVs-E4F262. E1A protein [E1A (289R)] was detected by polyclonal antibodies against the E1A(13S) product. (C) Regions of p50E4F required for stimulation of the E4 promoter. Relative E4-CAT activities were determined after cotransfection of 5 ng of pCMV-E1A(13S) and 30 ng of pCMVs constructs expressing E4F262 or E4F262 truncation mutants (N1, N2, N5, and C1). Values for each mutant were derived from two to four independent experiments. (D) Regions of p50E4F required for squelching of the E4 promoter. Relative E4-CAT activities were determined after cotransfection of 500 ng of pCMV-E1A(13S) and 500 ng of pCMVs constructs expressing E4F262 or E4F262 truncation mutants (N1, N2, N5, C1, C2, and C3). Values for each mutant were derived from two to six individual experiments. (E) Schematic depiction of p50E4F domains that are required for suppression of E1A-mediated transformation and regulation of the adenovirus E4 promoter.