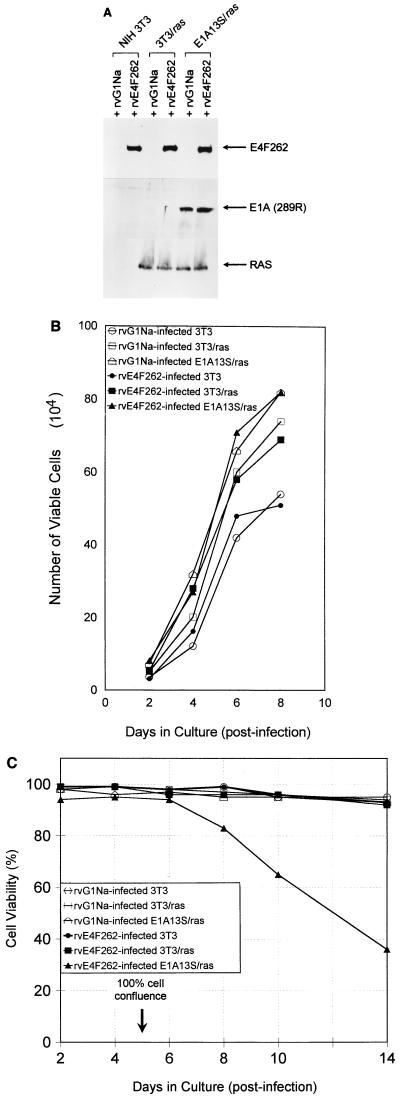
FIG. 5.
Retroviral expression of E4F262 reduces cell viability in confluent cultures of E1A13S/ras cells. (A) Western blots of lysates from NIH 3T3, 3T3/ras, and E1A13S/ras cells infected with rvE4F262 or control retrovirus rvG1Na. Polyclonal antibodies were used to detect E4F262 and E1A(289R) proteins in total cell lysates; rat monoclonal immunoglobulin G was used to detect Ras proteins in lysates of membrane-enriched fractions. Lysates were harvested 96 h postinfection. (B) NIH 3T3, 3T3/ras, and E1A13S/ras cells were infected with rvE4F262 or rvG1Na (MOI = 5 to 10 CFU/cell) for 24 h and grown in 35-mm-diameter wells after replating at 104 cells/well. Cell counts were performed after trypan blue staining at the indicated times. Growth curves were performed in triplicate, with a standard deviation of ≤6% between triplicates for all points. Cell viability was ≥95% at all points. NIH 3T3 cells were a single parental line; 3T3/ras cells were a pool of one Neor- and one Hygror NIH 3T3 cell lines that both express activated ras; E1A13S/ras cells were a pool of four Neor and Hygror NIH 3T3 cell lines that express the E1A(13S) cDNA and activated ras. (C) rvE4F262- or rvG1Na-infected NIH 3T3, 3T3/ras, and E1A13S/ras cells were replated in 17-mm-diameter wells at 2.0 × 104 cells/well and received fresh medium every 48 h. Cell viability was determined by trypan blue exclusion at the indicated times. Viability curves were performed in duplicate in two independent experiments, with a standard deviation of ≤9% between duplicates for all points in both experiments. Cell confluence was determined by visual inspection.