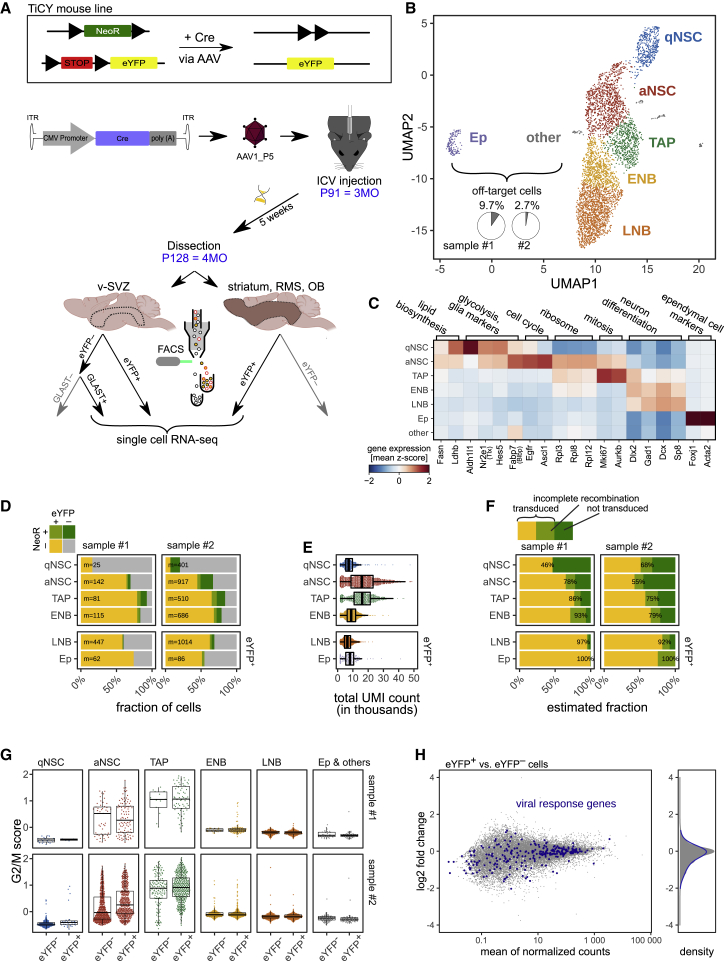
Figure 3.
Single-cell RNA sequencing (scRNA-seq) reveals transduction of cells of the adult NSC lineage by AAV1_P5
(A) Experimental outline of labeling, isolation, and scRNA-seq of the adult NSC lineage using the AAV1_P5 capsid. (Top panel) Untransduced cells from the TiCY mouse line express neomycin resistance (NeoR). Cre recombinase (Cre)-mediated recombination induces the expression of eYFP and the loss of NeoR expression. AAV1_P5 loaded with Cre was delivered to the lateral ventricle of P91 TiCY mice. After 5 weeks, all labeled (eYFP+) cells from the v-SVZ and the rest of the brain (striatum, rostral migratory stream [RMS], and OB), as well as further unlabeled NSC lineage cells (GLAST+ from v-SVZ) were sorted and used for scRNA-seq. (B) 2D representation of the resulting 4,572 single-cell transcriptomes. Most cells form one continuous trajectory from qNSCs to early NBs (ENBs; mostly from v-SVZ) and late NBs (LNBs)/immature neurons (mostly from rest of brain). Few off-target cells including Ep cells and others (gray) were captured. (C) Mean relative gene expression of NSC lineage markers from Llorens-Bobadilla et al.6 and Ep cell markers from Shah et al.58 in each cluster of single cells. (D) Fraction of eYFP+ and NeoR+ single-cell transcriptomes by cell type (m, cells per group). (E) Total number of uniquely identified mRNA molecules (UMI count) per cell, separated by cell type. (F) Maximum likelihood estimate of the fraction of transduced cells, based on values in (C and D). LNB and Ep were sorted by eYFP+ only and act as a control with an expected transduction rate of 100%. (G) Expression of G2/M-phase marker genes across samples and cell types (clusters from B), distinguishing between eYFP+ and eYFP− cells. (H, left) MA plot of gene-expression differences between eYFP+ and eYFP− cells. (Right) log2 fold-change distribution for all genes (gray) and viral response genes (blue).