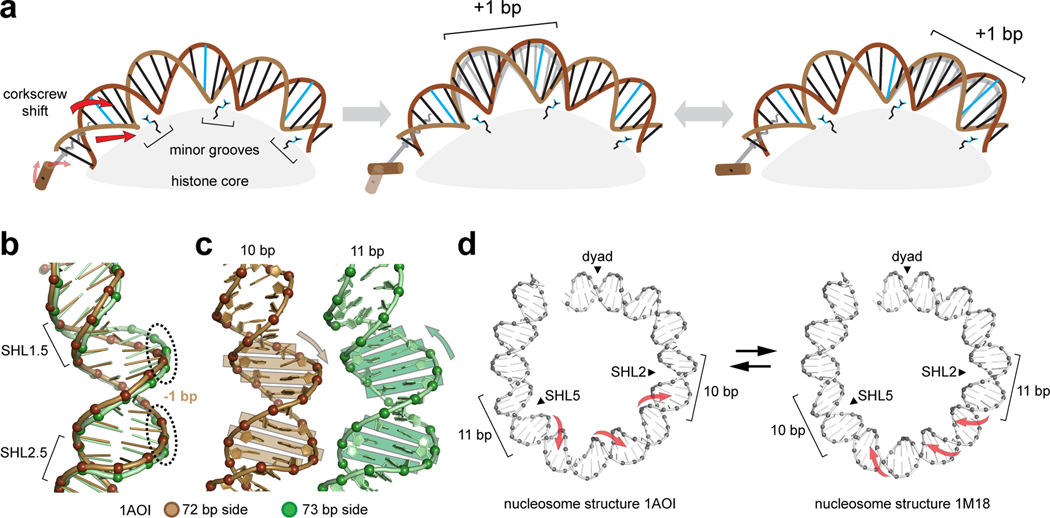
Figure 2. Twist defects on the nucleosome.
(a) The twist diffusion model for nucleosome repositioning (123, 124). A corkscrew shift of the duplex causes a DNA segment between two minor groove contacts to absorb an additional bp, which locally changes DNA twist (a twist defect). Once formed, a corkscrew shift on the other side transfers the twist defect further onto the nucleosome, accompanied by a 1 bp translation of DNA.
(b) Changes in DNA geometry accompanying twist defects. Comparison of the two sides of the original 146 bp nucleosome structure at SHL2 shows that the one bp difference is distributed ~5 nt away on the two strands, farthest from histone-DNA contacts.
(c) As highlighted by rectangular boxes, the shift in the DNA backbone at the farthest point from the histone core correlates with bp tilting.
(d) Two nucleosome crystal structures that captured twist defects at two separated locations, SHL2 and SHL5 (80, 117). These nucleosomes had the same DNA sequence and length, only differing by the presence or absence of a pyrrole-imidazole polyamide at SHL4 (1M18 structure). Red arrows represent expected corkscrew rotations of intervening DNA needed for a twist defect to collapse at one location and appear at the other.