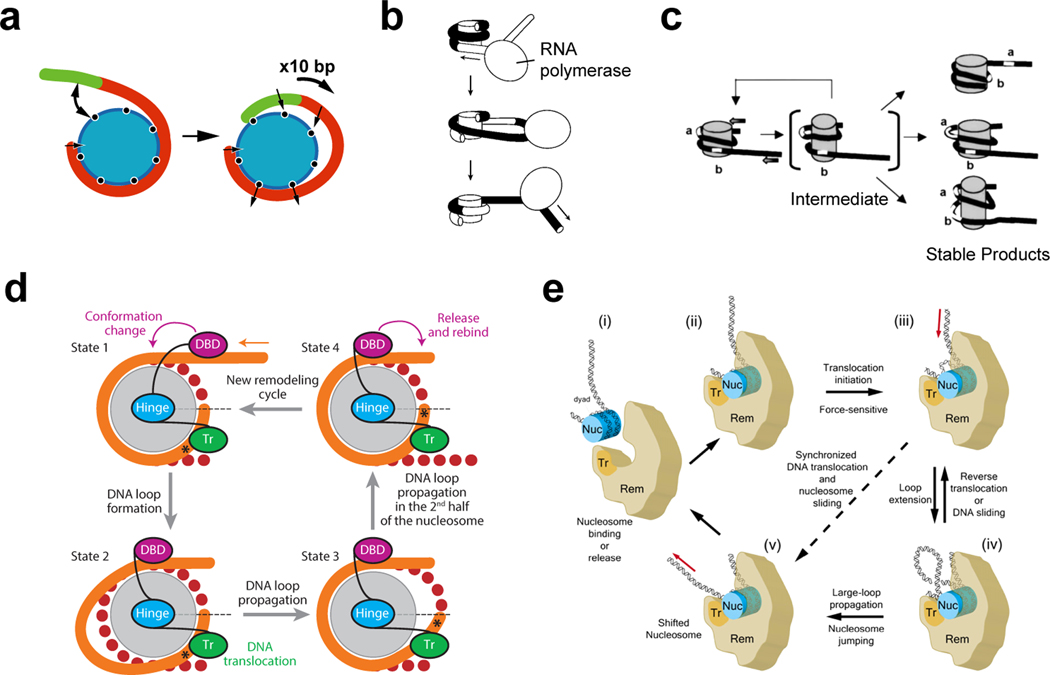
Figure 3. Bulge/loop propagation models of nucleosome sliding.
(a) A loop recapture model. In this model, DNA unwraps and then rewraps with a shifted position, trapping a loop of DNA. Adapted from Längst & Becker 2004 (67).
(b) Nucleosome repositioning resulting from transcription by an RNA polymerase around the histone core. Adapted with permission from Studitsky et al., 1997 (114).
(c) A model for how the SWI/SNF remodeler alters nucleosome structure, creating persistent DNA loops on the histone surface. Adapted from Fan et al., 2003 (37).
(d) A loop propagation model where the entry-side loop is created by cooperation between the ATPase motor at SHL2 (Tr) and a DNA-binding domain (DBD) at the nucleosome edge. DNA movement past the ATPase motor occurs between states 2 and 3, highlighted by the shift of an asterisk (*) reference point. Adapted from Clapier & Cairns, 2009 (25).
(e) A model where translocation of the RSC ATPase motor (Tr) at the internal SHL2 site directly spools out DNA into a loop at the dyad. Adapted from Zhang et al., 2006 (141).